CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
File:GH3 2013 Fig4.png
Jump to navigation
Jump to search
GH3_2013_Fig4.png (720 × 540 pixels, file size: 356 KB, MIME type: image/png)
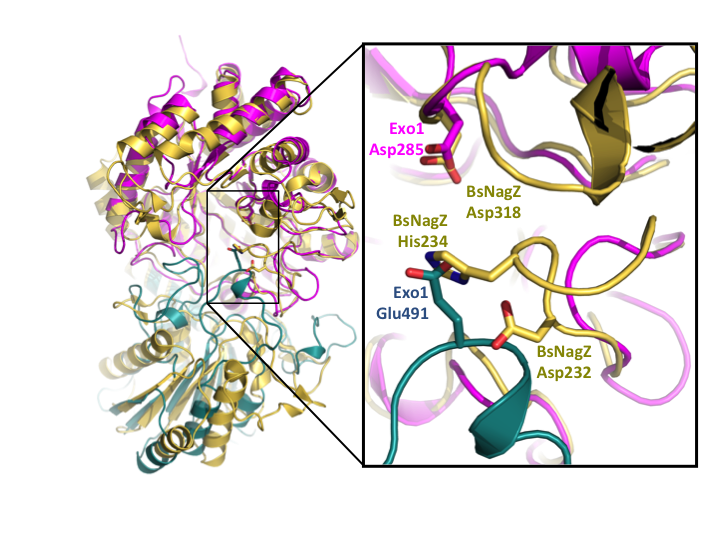
Figure 4. Overlay of barley β-glucan exohydrolase isoenzyme ExoI (domain 1 in magenta, and domain 2 in cyan) with the two-domain GH3 NagZ from B. subtilis (BsNagZ) (yellow) (PDB: 3BMX). GH3 NagZ enzymes contain a conserved histidine/aspartate dyad within a flexible loop of the catalytic domain that has been proposed as the general acid/base. In contrast to Exo1, the additional domain of BsNagZ does not participate in catalysis. The catalytic Asp nucleophile however, is conserved across the GH3 family, including the NagZ enzymes.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 12:36, 10 April 2013 |  | 720 × 540 (356 KB) | Brian Mark (talk | contribs) | '''Figure 4.''' '''Overlay of barley β-glucan exohydrolase isoenzyme ExoI (domain 1 in magenta, and domain 2 in cyan) with the two-domain GH3 NagZ from ''B. subtilis'' (BsNagZ) (yellow) (PDB: 3BMX). '''GH3 NagZ enzymes contain a conse... |
You cannot overwrite this file.
File usage
The following page uses this file: