CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Auxiliary Activity Family 9"
Paul Harris (talk | contribs) |
Harry Brumer (talk | contribs) |
||
| Line 28: | Line 28: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | This family was originally placed among the glycoside hydrolases in family [[GH61]] based largely on a report of very weak endoglucanase activity of “Cel61A” from ''Trichoderma reesei'' using substrates such as carboxymethyl cellulose and ß-glucan <cite>Karlsson2001</cite>. Subsequently, other investigations seemed to confirm this result with other AA9 proteins (e.g.<cite>Bauer2006</cite>). It remains unclear whether the activities observed could be ascribed to AA9 or to low levels of contaminating proteins. It is now believed that AA9 proteins are not endoglucanases or glycoside hydrolases at all but rather lytic polysaccharide monooxygenases (LPMOs) that cleave polysaccharides by a novel oxidative mechanism (see below). Current published evidence indicates that cellulose is the preferred substrate, however many AA9 proteins show little or no activity with cellulose and may have other substrate specificities, or possibly very different co-factor requirements. This is a very large family and only a fraction of its members have been assayed for activity on cellulose and an even smaller number assayed on other abundant polysaccharide substrates such as chitin, starch and xylan. Given the structural and mechanistic similarities between AA9 and chitin-active [[AA10]] (formerly [[CBM33]])and AA11 proteins, and the ability of some [[AA10]] proteins to cleave cellulose as well as chitin, it would not be surprising if some members of the AA9 family had substrate specificities other than cellulose. It is also not clear that polysaccharides would be the only potential substrates, and thus the term LPMO would prudently be reserved for those AA9 family members with proven lytic activity on polysaccharides. | + | This family was originally placed among the glycoside hydrolases in family [[GH61]] based largely on a report of very weak endoglucanase activity of “Cel61A” from ''Trichoderma reesei'' using substrates such as carboxymethyl cellulose and ß-glucan <cite>Karlsson2001</cite>. Subsequently, other investigations seemed to confirm this result with other AA9 proteins (e.g.<cite>Bauer2006</cite>). It remains unclear whether the activities observed could be ascribed to AA9 or to low levels of contaminating proteins. It is now believed that AA9 proteins are not endoglucanases or glycoside hydrolases at all but rather lytic polysaccharide monooxygenases (LPMOs) that cleave polysaccharides by a novel oxidative mechanism (see below). Current published evidence indicates that cellulose is the preferred substrate, however many AA9 proteins show little or no activity with cellulose and may have other substrate specificities, or possibly very different co-factor requirements. This is a very large family and only a fraction of its members have been assayed for activity on cellulose and an even smaller number assayed on other abundant polysaccharide substrates such as chitin, starch and xylan. Given the structural and mechanistic similarities between AA9 and chitin-active [[AA10]] (formerly [[CBM33]]) and AA11 proteins, and the ability of some [[AA10]] proteins to cleave cellulose as well as chitin, it would not be surprising if some members of the AA9 family had substrate specificities other than cellulose. It is also not clear that polysaccharides would be the only potential substrates, and thus the term LPMO would prudently be reserved for those AA9 family members with proven lytic activity on polysaccharides. |
Current thinking posits that AA9 and [[AA10]] proteins act on relatively crystalline surface regions of their substrates and thereby create attachment sites and enhanced accessibility for the canonical glycoside hydrolases that subsequently further hydrolyze the substrate. This hypothesis has not yet been rigorously tested but is the most parsimonious explanation for the ability of these proteins to enhance glycoside hydrolase action. The relatively flat active-site surface of these proteins is consistent with binding to an ordered polysaccharide surface, although the mechanism of binding appears to involve mostly H-bonding interactions in the case of [[AA10]] <cite>Aachmann2012 Vaaje-Kolstad2012</cite> versus stacking with planar aromatic residues for AA9 <cite>Li2012 Wu2013</cite>. Soluble polysaccharides have not been shown to be a substrate for any AA9 proteins except for one from Neurospora crassa <cite>Isaksen2013</cite>. | Current thinking posits that AA9 and [[AA10]] proteins act on relatively crystalline surface regions of their substrates and thereby create attachment sites and enhanced accessibility for the canonical glycoside hydrolases that subsequently further hydrolyze the substrate. This hypothesis has not yet been rigorously tested but is the most parsimonious explanation for the ability of these proteins to enhance glycoside hydrolase action. The relatively flat active-site surface of these proteins is consistent with binding to an ordered polysaccharide surface, although the mechanism of binding appears to involve mostly H-bonding interactions in the case of [[AA10]] <cite>Aachmann2012 Vaaje-Kolstad2012</cite> versus stacking with planar aromatic residues for AA9 <cite>Li2012 Wu2013</cite>. Soluble polysaccharides have not been shown to be a substrate for any AA9 proteins except for one from Neurospora crassa <cite>Isaksen2013</cite>. | ||
Revision as of 13:34, 9 January 2014
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Paul Harris^^^
- Responsible Curator: ^^^Paul Harris^^^
| Auxiliary Activity Family AA9 | |
| Clan | Structurally related to AA10 & AA11 |
| Mechanism | lytic oxidase |
| Active site residues | mononuclear copper ion |
| CAZy DB link | |
| http://www.cazy.org/AA9.html | |
Substrate specificities
This family was originally placed among the glycoside hydrolases in family GH61 based largely on a report of very weak endoglucanase activity of “Cel61A” from Trichoderma reesei using substrates such as carboxymethyl cellulose and ß-glucan [1]. Subsequently, other investigations seemed to confirm this result with other AA9 proteins (e.g.[2]). It remains unclear whether the activities observed could be ascribed to AA9 or to low levels of contaminating proteins. It is now believed that AA9 proteins are not endoglucanases or glycoside hydrolases at all but rather lytic polysaccharide monooxygenases (LPMOs) that cleave polysaccharides by a novel oxidative mechanism (see below). Current published evidence indicates that cellulose is the preferred substrate, however many AA9 proteins show little or no activity with cellulose and may have other substrate specificities, or possibly very different co-factor requirements. This is a very large family and only a fraction of its members have been assayed for activity on cellulose and an even smaller number assayed on other abundant polysaccharide substrates such as chitin, starch and xylan. Given the structural and mechanistic similarities between AA9 and chitin-active AA10 (formerly CBM33) and AA11 proteins, and the ability of some AA10 proteins to cleave cellulose as well as chitin, it would not be surprising if some members of the AA9 family had substrate specificities other than cellulose. It is also not clear that polysaccharides would be the only potential substrates, and thus the term LPMO would prudently be reserved for those AA9 family members with proven lytic activity on polysaccharides.
Current thinking posits that AA9 and AA10 proteins act on relatively crystalline surface regions of their substrates and thereby create attachment sites and enhanced accessibility for the canonical glycoside hydrolases that subsequently further hydrolyze the substrate. This hypothesis has not yet been rigorously tested but is the most parsimonious explanation for the ability of these proteins to enhance glycoside hydrolase action. The relatively flat active-site surface of these proteins is consistent with binding to an ordered polysaccharide surface, although the mechanism of binding appears to involve mostly H-bonding interactions in the case of AA10 [3, 4] versus stacking with planar aromatic residues for AA9 [5, 6]. Soluble polysaccharides have not been shown to be a substrate for any AA9 proteins except for one from Neurospora crassa [7].
Kinetics and Mechanism
It was known for many years from the patent literature (e.g. US20060005279, US20070077630, WO2005074647) and conference proceedings that some AA9 proteins could dramatically enhance the activity of canonical cellulase mixtures in the hydrolysis of lignocellulose, however this information was not published in a peer-reviewed journal until 2010 [8]. An explanation for this enhancement remained elusive for many years, but a first clue was the demonstration in the 2010 publication that metal ion is required for the enhancement and that the enhancement is not evident with relatively pure cellulose (Avicel) but only with pretreated lignocellulose such as acid-pretreated corn stover. In hindsight it became clear that the complex lignocellulosic substrate was providing an essential cofactor(s) that the Avicel did not. Subsequent studies, again first published in the patent literature (US20100159536) and subsequently in journals [9, 10], showed that a combination of AA9 and cellobiose dehydrogenase could cleave pure cellulose. Additional work showed that a large number of small redox-active molecules such as gallate, ascorbate and catechol could replace cellobiose dehydrogenase in this regard, and both soluble and insoluble lignin and lignin derivatives might also act in this capacity [11]. However, the “natural” redox-active cofactors for AA9 proteins are currently unknown and may well vary depending on availability and the specific AA9 protein. Cellobiose dehydrogenase is often mentioned as a potential natural cofactor and indeed is often co-secreted with AA9 proteins. However there are many species of filamentous fungi (e.g. Paxillus involutus, Talaromyces stipitatus, Talaromyces thermophilus, and several Trichoderma (Hypocrea) species) that do not produce cellobiose dehydrogenase (based on lack of an obvious gene encoding it) and yet have multiple AA9-encoding genes. Nonetheless, for those fungi that do secrete cellobiose dehydrogenase, one of its main functions may be to synergize with AA9 proteins, presumably by acting as an electron donor.
A key publication in 2010 [12] showed that AA10, a structural relative of AA9, could oxidatively degrade crystalline chitin in the presence of metal ion and the reductant ascorbate, producing a mixture of oxidized and unoxidized oligosaccharides. Shortly thereafter, similar data were published on AA9 [13, 14, 15] (reviewed by [16, 17, 18, 19]) showing that these proteins are copper-dependent monooxygenases which are capable of inserting oxygen at the C-1 or C-4 (and perhaps C-6) position of the glycosidic bond in cellulose. The so-called type 1 LPMOs produce predominantly oxidation at C-1 (reducing end), probably initially producing an aldonolactone that spontaneously hydrolyzes to an aldonic acid. Type 2 LPMOs generate predominantly C-4 non-reducing end oxidized products (4-ketoaldolase and the hydrated gemdiol), and type 3 LPMOs appear to produce oxidized products at both the reducing and nonreducing ends [7, 10, 13, 15]. Results consistent with C-6 oxidation have also been reported [20]. Positioning of the substrate on the catalytic surface very likely determines the site of oxidation [5]. The detailed interactions between AA9, bound metal ion, redox-active cofactor and substrate remain an area of active investigation, and additional surprises are likely. At present, the ability of AA9 proteins to oxidize cellulose is thought to arise from generation of oxidizing copper-oxygen species (superoxides or peroxides) at the active site by a mechanism that remains unclear (reviewed by Hemsworth et al. [16]). Presumably Cu(I) at the active site activates a molecular oxygen and the activated oxygen species becomes incorporated into the cellulose chain. A reductant is then necessary to regenerate Cu(I) [5, 15]. Quantum mechanical calculations favor a copper-oxyl radical mechanism for hydrogen abstraction and subsequent hydroxylation [21].
Catalytic Residues
AA9 proteins are quite diverse in sequence with identities between some sequences below 30% within the main catalytic domain. However, the catalytic copper ion is coordinated by a highly conserved “histidine brace” consisting of the imidazole and backbone amino group of the N-terminal histidine and the imidazole of an additional histidine. Closely associated with the catalytic copper in most AA9 proteins is the phenolic oxygen of a highly conserved tyrosine that could play a redox role in catalysis since mutation to phenylalanine greatly reduces but does not entirely eliminate activity in “GH61E” from Thielavia terrestris [8]. About 3% of all sequenced AA9 proteins and the majority of AA10 proteins naturally contain phenylalanine at this position rather than tyrosine. Additional contributions to catalysis might come from long distance internal electron transfer through a conserved ionic network and conserved aromatic residues [5], although this has not yet been directly demonstrated.
Three-dimensional structures
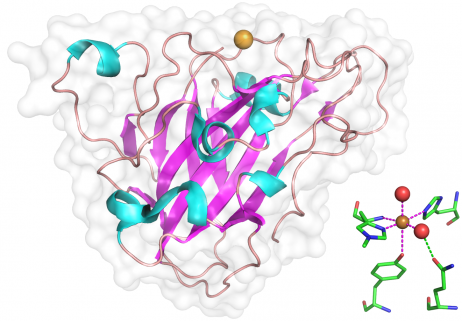
In the structures so far determined [5, 6, 8, 13, 22], the core of the catalytic domain adopts an immunoglobulin-like ß-sandwich fold with the individual ß-strands connected by long loops that have one to several helical insertions of variable length. Families AA9 and AA10 share a common ß-sandwich fold and a similarly flat surface that contains the catalytic metal binding site. Different metal ions have been observed in different structures depending on which metal was most abundantly present in the crystallization buffer, however copper appears to be the "native" metal ion. In structures of AA9 published to date, the N-terminal histidine is methylated at Nε. The function of this methylation, if any, is unknown, but could serve to increase pKa of the histidine or to modulate redox activity. To date only the structures of the core catalytic domains have been solved. Many AA9 proteins have conserved C-terminal extensions beyond the obvious catalytic domain and these have no obvious identity to anything outside of the AA9 family (e.g. Uniprot N4XCE3). Others have recognizable linker domains with a family 1 cellulose binding module (CBM1) or other conserved modules of unknown function.
Family Firsts
- First AA9 protein identified
- CelI from Agaricus bisporus was the first reported in the literature [23]. It was induced by growth on cellulose, but no function was assigned later to the heterologously expressed protein [24, 25].
- First demonstration of synergy between AA9 and canonical glycoside hydrolases
- Experiments performed in 2003 by Novozymes, Inc. demonstrated the ability of AA9 proteins to lower the dose of cellulase required to reach a given level of cellulose hydrolysis. These results were first reported in technical reports to the National Renewable Energy Laboratory and US DOE and in patent applications (e.g. US20060005279 A1), subsequently at conferences and in a book review chapter [26], and more completely in 2010 [8].
- First demonstration of oxidative cleavage by an AA9 protein
- The ability of an AA9 protein from Thermoascus aurantiacus (“GH61A”) to degrade cellulose in the presence of cellobiose dehydrogenase was shown first in patent application US20100159536. Experimental details were subsequently published [9], followed closely thereafter by work showing that the same protein was very likely a copper-dependent oxidase [13]. Related work showing the oxidative action of family AA10 on chitin was published by the Eijsink lab in 2010 [12].
- First 3-D structure
- The structure of “Cel61B” from Trichoderma reesei (Hypocrea jecorina) was the first published in peer-reviewed literature [22].
References
- Karlsson J, Saloheimo M, Siika-Aho M, Tenkanen M, Penttilä M, and Tjerneld F. (2001). Homologous expression and characterization of Cel61A (EG IV) of Trichoderma reesei. Eur J Biochem. 2001;268(24):6498-507. DOI:10.1046/j.0014-2956.2001.02605.x |
- Bauer S, Vasu P, Persson S, Mort AJ, and Somerville CR. (2006). Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci U S A. 2006;103(30):11417-22. DOI:10.1073/pnas.0604632103 |
- Aachmann FL, Sørlie M, Skjåk-Bræk G, Eijsink VG, and Vaaje-Kolstad G. (2012). NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc Natl Acad Sci U S A. 2012;109(46):18779-84. DOI:10.1073/pnas.1208822109 |
- Vaaje-Kolstad G, Bøhle LA, Gåseidnes S, Dalhus B, Bjørås M, Mathiesen G, and Eijsink VG. (2012). Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J Mol Biol. 2012;416(2):239-54. DOI:10.1016/j.jmb.2011.12.033 |
- Li X, Beeson WT 4th, Phillips CM, Marletta MA, and Cate JH. (2012). Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure. 2012;20(6):1051-61. DOI:10.1016/j.str.2012.04.002 |
- Wu M, Beckham GT, Larsson AM, Ishida T, Kim S, Payne CM, Himmel ME, Crowley MF, Horn SJ, Westereng B, Igarashi K, Samejima M, Ståhlberg J, Eijsink VG, and Sandgren M. (2013). Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the Basidiomycota fungus Phanerochaete chrysosporium. J Biol Chem. 2013;288(18):12828-39. DOI:10.1074/jbc.M113.459396 |
- Isaksen T, Westereng B, Aachmann FL, Agger JW, Kracher D, Kittl R, Ludwig R, Haltrich D, Eijsink VG, and Horn SJ. (2014). A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2014;289(5):2632-42. DOI:10.1074/jbc.M113.530196 |
- Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, and Lo Leggio L. (2010). Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49(15):3305-16. DOI:10.1021/bi100009p |
- Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, and Sweeney MD. (2011). Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol. 2011;77(19):7007-15. DOI:10.1128/AEM.05815-11 |
- Phillips CM, Beeson WT, Cate JH, and Marletta MA. (2011). Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6(12):1399-406. DOI:10.1021/cb200351y |
- Dimarogona M, Topakas E, Olsson L, and Christakopoulos P. (2012). Lignin boosts the cellulase performance of a GH-61 enzyme from Sporotrichum thermophile. Bioresour Technol. 2012;110:480-7. DOI:10.1016/j.biortech.2012.01.116 |
- Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, and Eijsink VG. (2010). An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219-22. DOI:10.1126/science.1192231 |
- Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen JC, Johansen KS, Krogh KB, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, and Walton PH. (2011). Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A. 2011;108(37):15079-84. DOI:10.1073/pnas.1105776108 |
- Westereng B, Ishida T, Vaaje-Kolstad G, Wu M, Eijsink VG, Igarashi K, Samejima M, Ståhlberg J, Horn SJ, and Sandgren M. (2011). The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS One. 2011;6(11):e27807. DOI:10.1371/journal.pone.0027807 |
- Beeson WT, Phillips CM, Cate JH, and Marletta MA. (2012). Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134(2):890-2. DOI:10.1021/ja210657t |
- Hemsworth GR, Davies GJ, and Walton PH. (2013). Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660-8. DOI:10.1016/j.sbi.2013.05.006 |
- Dimarogona M, Topakas E, and Christakopoulos P. (2013). Recalcitrant polysaccharide degradation by novel oxidative biocatalysts. Appl Microbiol Biotechnol. 2013;97(19):8455-65. DOI:10.1007/s00253-013-5197-y |
- Horn SJ, Vaaje-Kolstad G, Westereng B, and Eijsink VG. (2012). Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(1):45. DOI:10.1186/1754-6834-5-45 |
-
Lo Leggio, L, Welner, DH, and De Maria, L. A structural overview of GH61 proteins - fungal cellulose degrading polysaccharide monooxygenases. Computational and Structural Biotechnology Journal. 2012 Sep;2(3).
- Bey M, Zhou S, Poidevin L, Henrissat B, Coutinho PM, Berrin JG, and Sigoillot JC. (2013). Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl Environ Microbiol. 2013;79(2):488-96. DOI:10.1128/AEM.02942-12 |
- Kim S, Ståhlberg J, Sandgren M, Paton RS, and Beckham GT. (2014). Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc Natl Acad Sci U S A. 2014;111(1):149-54. DOI:10.1073/pnas.1316609111 |
- Karkehabadi S, Hansson H, Kim S, Piens K, Mitchinson C, and Sandgren M. (2008). The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J Mol Biol. 2008;383(1):144-54. DOI:10.1016/j.jmb.2008.08.016 |
- Raguz S, Yagüe E, Wood DA, and Thurston CF. (1992). Isolation and characterization of a cellulose-growth-specific gene from Agaricus bisporus. Gene. 1992;119(2):183-90. DOI:10.1016/0378-1119(92)90270-y |
- Armesilla AL, Thurston CF, and Yagüe E. (1994). CEL1: a novel cellulose binding protein secreted by Agaricus bisporus during growth on crystalline cellulose. FEMS Microbiol Lett. 1994;116(3):293-9. DOI:10.1111/j.1574-6968.1994.tb06718.x |
- Yagüe E, Wood DA, and Thurston CF. (1994). Regulation of transcription of the cel1 gene in Agaricus bisporus. Mol Microbiol. 1994;12(1):41-7. DOI:10.1111/j.1365-2958.1994.tb00993.x |
- Merino ST and Cherry J. (2007). Progress and challenges in enzyme development for biomass utilization. Adv Biochem Eng Biotechnol. 2007;108:95-120. DOI:10.1007/10_2007_066 |