CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Catalytic nucleophile
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
Overview
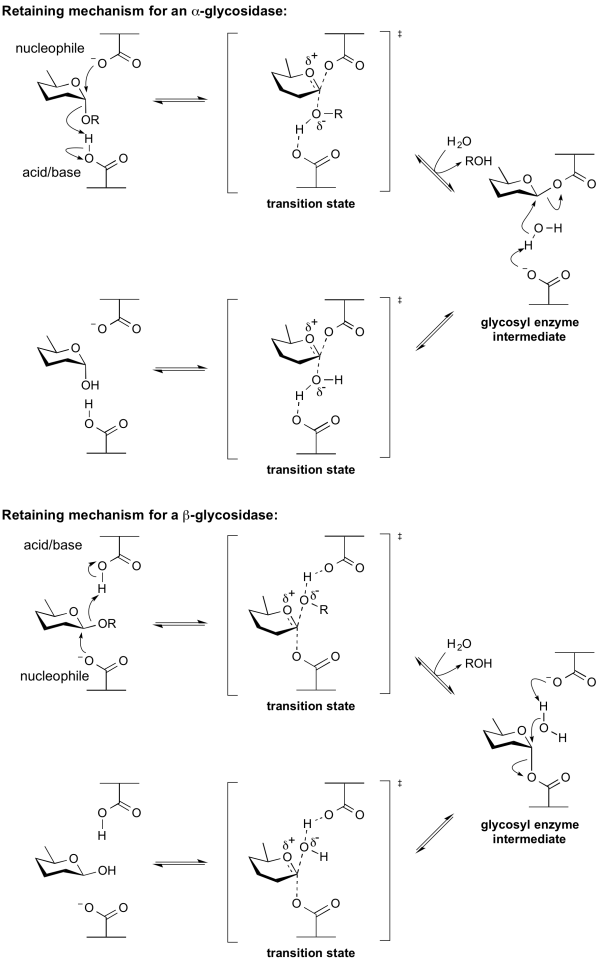
The term catalytic nucleophile refers to an amino acid residue in a glycoside hydrolase. The residue participates in the classical Koshland retaining mechanism. In summary, hydrolysis occurs with net retention of configuration through a two step, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. The reaction occurs with acid/base assistance provided by another amino acid side chain. In the first step (often called the glycosylation step), the catalytic nucleophile plays the role of a nucleophile, attacking the anomeric centre to displace the aglycon and form a glycosyl enzyme intermediate. At the same time the other residue functions as an acid catalyst and protonates the glycosidic oxygen as the bond cleaves. In the second step (known as the deglycosylation step), the glycosyl enzyme is hydrolyzed by water, with the other residue now acting as a base catalyst deprotonating the water molecule as it attacks. At the completion of the catalytic cycle the catalytic nucleophile is restored.
Methods for identifying the catalytic nucleophile
The main methods for assigning a particular amino acid with the function of a catalytic nucleophile is either by labelling with a mechanism-based inhibitor, or through kinetic analysis of mutants. In the absence of these data, X-ray crystallography, especially of complexes with substrate or transition state analogues, may provide clues towards the identity of the catalytic nucleophile.
Kinetic analysis of mutants
Assignment of the catalytic nucleophile may be achieved by the kinetic analysis of enzyme mutants in which the candidate residues have been replaced by a non-nucleophilic residue. Alanine is preferred as a replacement residue owing to its small size. Glycine is non-optimal, as the lack of an alpha-substituent may lead to differences in conformation. Based on empirical observations, nucleophile mutants typically give kcat values 10,000-fold lower than the wildtype enzyme [1]. A second kinetic test for the nature of the nucleophile is the observation of a 'chemical rescue'. When supplied with a glycoside with a good leaving group, such as 2,4-dintrophenyl, such 'activated' glycosides may exhibit an increased rate of turnover in the presence of small anionic nucleophiles such as azide, fluoride or formate. In this case the product of 'chemical rescue' should be a glycosyl azide, fluoride or formate with inverted anomeric stereochemistry.
Mechanism-based labelling
When treated with a 'mechanism-based' inhibitor, the catalytic nucleophile may be covalently labelled with the turnover product. The location of the covalently attached group may be determined by X-ray crystallography, or peptide mapping (commonly using MS/MS techniques). The two main approaches used for mechanism-based labelling of the catalytic nucleophile rely on the use of modified substrates in which the deglycosylation rate has been selectively reduced, or through the use of enzyme mutants in concert with substrates in which deglycosylation is rate limiting.
2-Deoxy-2-fluoro- and 5-fluoro sugars bearing good leaving groups (eg fluoride or 2,4-dinitrophenyl) are good candidates for covalent modification of the catalytic nucleophile [2, 3]. In these compounds the presence of the 2- or 5-fluorine substituents leads to destabilization of oxocarbenium ion-like transition states, which alone would lead to rate reductions in the glycosylation and deglycosylation steps. However, the incorporation of a good leaving group (fluoride or 2,4-dinitrophenolate) leads to a selective acceleration of the glycosylation step relative to the deglycosylation step, and this potentially allows accumulation of the glycosyl enzyme intermediate. The glycosyl enzyme intermediate may have a stability with a half-life on the order of seconds to days, allowing the use of X-ray crystallography or peptide mapping studies with MS/MS. As the labelling of the enzyme is mechanism-based, it is argued that this technique provides high selectivity for labelling of the catalytic nucleophile. Most notably this technique has been applied to the labelling of the catalytic nucleophile of the 'textbook' enzyme hen egg-whie lysozyme [4].
The use of other electrophilic reagents, such as epoxyalkyl glycosides and various conduritol epoxides, has not proven to be as reliable, as the higher reactivity of these compounds has on some occasions led to the labelling of residues that have subsequently been proven not to be the catalytic nucleophile.
An alternative approach that has been used for some transglycosidases is the use of substrates that have been modified to remove the hydroxyl group that acts as the acceptor for the transglycosylation event. By use of a 'donor' substrate possessing a good leaving group aglycon but lacking the hydroxyl group that acts as the acceptor, the glycosyl enzyme intermediate can accumulate sufficiently so as to allow its study by X-ray crystallography and mass spectrometry.
Mutagenesis leading to intermediate accumulation
The glycosyl enzyme intermediate may also be trapped through the generation of an enzyme mutant that suffers a reduced rate of deglycosylation, thereby leading to accumulation of the intermediate [5]. Mutation of the acid/base residue leads to a reduction in the rate of both glycosylation and deglycosylation. By using a substrate possessing a good leaving group that does not require general acid catalysis, the glycosylation step is accelerated. On the other hand the deglyclosylation step requires general base catalysis and remains retarded, leading to accumulation of the glycosyl enzyme, which may be studied using the techniques outlined above.
References
- Ly HD and Withers SG. (1999). Mutagenesis of glycosidases. Annu Rev Biochem. 1999;68:487-522. DOI:10.1146/annurev.biochem.68.1.487 |
- Williams SJ and Withers SG. (2000). Glycosyl fluorides in enzymatic reactions. Carbohydr Res. 2000;327(1-2):27-46. DOI:10.1016/s0008-6215(00)00041-0 |
- Zechel DL and Withers SG. (2000). Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc Chem Res. 2000;33(1):11-8. DOI:10.1021/ar970172+ |
- Vocadlo DJ, Davies GJ, Laine R, and Withers SG. (2001). Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001;412(6849):835-8. DOI:10.1038/35090602 |
- Mosi R, He S, Uitdehaag J, Dijkstra BW, and Withers SG. (1997). Trapping and characterization of the reaction intermediate in cyclodextrin glycosyltransferase by use of activated substrates and a mutant enzyme. Biochemistry. 1997;36(32):9927-34. DOI:10.1021/bi970618u |