CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
File:GH3 2013 Fig6.png
GH3_2013_Fig6.png (720 × 540 pixels, file size: 245 KB, MIME type: image/png)
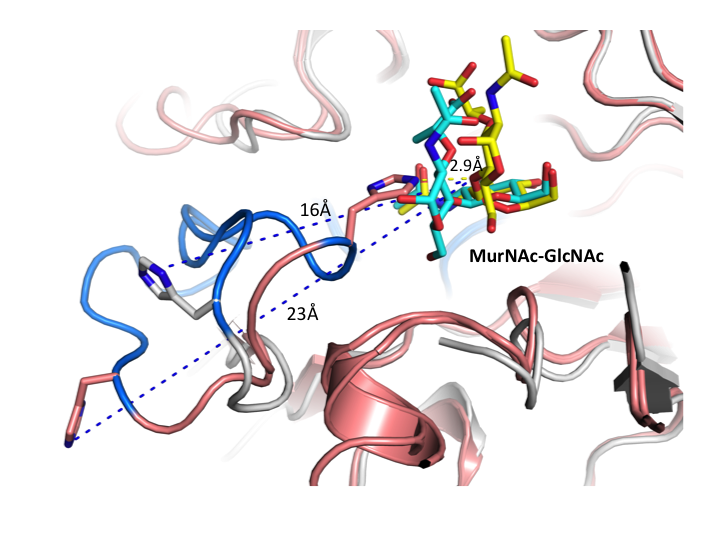
Figure 6. The conserved loop containing the proposed catalytic acid/base histidine of GH3 NagZ enzymes is highly mobile, which appears to drive substrate distortion to promote glycosidic bond hydrolysis[1]. Colour scheme: B. subtilis NagZ (BsNagZ) (yellow) (PDB: 4GYJ and 4GYK), S. typhimurium NagZ (StNagZ) (grey)(PDB: 4GVF).
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 12:44, 10 April 2013 |  | 720 × 540 (245 KB) | Brian Mark (talk | contribs) | '''Figure 6.''' '''The conserved loop containing the proposed catalytic acid/base histidine of GH3 NagZ enzymes is highly mobile, which''' '''appears to drive substrate distortion to promote glycosidic bond hydrolysis'''<cite>Bacik2012</cite>. Colo... |
You cannot overwrite this file.
File usage
The following page uses this file: