CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Auxiliary Activity Family 5
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Maria Cleveland^^^ and ^^^Yann Mathieu^^^
- Responsible Curator: ^^^Harry Brumer^^^
| Auxiliary Activity Family AA5 | |
| Fold | Seven-bladed β-propeller |
| Mechanism | Copper Radical Oxidase |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/AA5.html | |
Substrate Specificities
Enzymes from Auxiliary Activity Family 5 (AA5) are mononuclear copper-radical oxidases (CROs) that perform the two-electron oxidation of substrates using oxygen as the final electron acceptor (EC 1.1.3.-) [1]. AA5 members are further classified into two major subfamilies [2]: Subfamily AA5_1 contains characterized glyoxal oxidases (EC 1.2.3.15) [3]. Subfamily AA5_2 contains galactose 6-oxidases (EC 1.1.3.9), which oxidize the C-6 hydroxyl of diverse galactosides to the corresponding aldehyde [4, 5, 6]. AA5_2 also contains the more recently discovered general alcohol oxidases (EC 1.1.3.13) [6, 7, 8] and aryl alcohol oxidases (EC 1.1.3.7) [9, 10]. The first biochemically characterized member of AA5 was the galactose 6-oxidase from the phytopathogenic fungus Fusarium graminearum (previously known as Polyporus circinatus and Cladobotryum (Dactylium) dendroides [11]), which was originally reported in 1959 following isolation from cultures [12, 13]. Subsequently, the Fusarium graminearum galactose 6-oxidase became the defining member of AA5_2 [2]. The first characterized member of what is now known as AA5_1 is the glyoxal oxidase from Phanerochaete chrysosporium, which was likewise isolated from fungal culture [14].
In contrast to their fungal and bacterial counterparts, plant AA5 members do not fall within the two defined subfamilies. An AA5 enzyme from Arabidopsis thaliana has been demonstrated in vivo to have galactose 6-oxidase activity and promote cell-to-cell adhesion in the seed coat epidermis [15] (see also [16]. Additionally, a Streptomyces lividans enzyme, GlxA, which is distantly related to AA5, has been shown to oxidize glycolaldehyde and a deletion mutant showed a loss of glycan accumulation at hyphal tips [17].
AA5_1
The AA5_1 members are generally known as glyoxal oxidases (EC 1.2.3.15), characterized members of which typically accept a range of simple aldehydes, α-hydroxycarbonyl, and α-dicarbonyl compounds as substrates, with the highest activities observed on glyoxal, methylglyoxal and glycolaldehyde [1, 14, 18, 19, 20]. In contrast, two glyoxal oxidases form Pycnoporus cinnabarinus demonstrated the highest catalytic efficiency on glyoxylic acid [21]. An apparent distinction between the AA5_1 and AA5_2 subfamilies is that while AA5_1 enzymes catalyze the oxidation of aldehydes to carboxylic acids [1], AA5_2 members oxidize primary alcohols to the corresponding aldehyde (and, in some instances, also oxidize the aldehyde to the acid, albeit much more slowly) [4, 22]. Consequently, the oxidation of aldehydes by AA5 CROs has been suggested to proceed through the hydrated, gem-diol species [18].
AA5_2
The archetypal CRO and AA5 member is the Fusarium graminearum galactose 6-oxidase (FgrGalOx), which catalyzes the regioselective oxidation of the C6-hydroxyl group on the monosaccharide galactose (EC 1.3.3.7) [12, 13]. The range of substrates oxidized by FgrGalOx also includes galactosides as methyl beta-galactopyranoside [23], and galactose-containing di-, oligo-, and polysaccharides, including lactose, melibiose, raffinose, galactoxyloglucan, galactomannan and galactoglucomannan [22, 24]. Several other AA5_2 members from Fusarium species, such as those from F. oxysporum [25], F. sambucinum [23], and F. acuminatum [26] have substrate specificities similar to FgrGalOx. The last distinct group of AA5_2 enzymes based on substrate specificity are the raffinose-specific oxidases (EC 1.3.3.-). Other AA5_2 orthologs exhibit specificity for the alpha-galactosyl unit of the di- and trisaccharides mellibiose and raffinose, respectively, over galactose [5, 6]. For many decades since their discovery, galactose 6-oxidase activity was thought to be the defining feature of this family, although a limited ability of FgrGalOx to oxidize non-carbohydrate alcohols has been noted [27, 28]
In 2015, two AA5_2 orthologs from the fungi Colletotrichum graminicola and Colletotrichum gloeosporioides were characterized (CgrAlcOx and CglAlcOx, respectively), which were essentially inactive on galactose and galactosides, but efficiently oxidized the hydroxyl group of diverse aliphatic and aromatic primary alcohols [7]. These enzymes exhibited high catalytic efficiency towards, e.g., n-butan-1-ol, 2,4-hexadiene-1-ol, benzyl alcohol, and cinnamyl alcohol, and were therefore denoted as general alcohol oxidases (EC 1.3.3.13) [7]. Likewise, two AA5_2 members were characterized from the pathogenic fungi Pyricularia oryzae (PorAlcOx) and Colletotrichum higginsianum (ChiAlcOx), which exhibited prominent activity on n-butan-1-ol, ethanol, 1,3-butanediol, and glycerol [8]. Since then, additional AA5_2 enzymes from various fungi have been characterized as general alcohol oxidases, some of which efficiently oxidize both carbohydrate and non-carbohydrate substrates [6]. More specifically, several fungal AA5_2 members, including homologs from Colletotrichum/Glomerella and Fusarium species, have been characterized as aryl alcohol oxidases due to predominant specificities toward substituted benzyl alcohols and 5-hydroxymethylfurfural (HMF) (see EC 1.1.3.7 and EC 1.1.3.47) [6, 9, 10].
The specificity of AA5 CROs has been harnessed for a range of biotechnological applications. The earliest examples include glycoprotein labelling via oxidation of galactosyl residues with FgrGalOx [ADD REFS]. Likewise, FgrGalOx has been utilized for the production of functionalized carbohydrates from biomass sources [29, 30, 31, 32, 33, 34, 35, 36]. The ability of specific AA5 members to oxidize aliphatic and aromatic alcohols to the corresponding aldehydes, including stereoselectively, has biocatalytic applications in chemical production, e.g. for the pharmaceutical and food and fragrance industries [ADD REFS] [37, 38, 39]. Similarly, the ability of CROs to convert HMF into the bi-functional precursors diformylfuran (DFF) and 2,5-furandicarboxylic acid (FDCA) may find application in polymer manufacturing [9] [ADD REFS]. Several variants of FgrGalOx have been developed to enable such applications [ADD REFS].
Kinetics and Mechanism
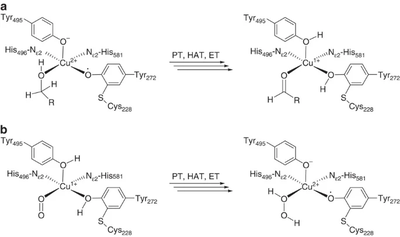
The majority of what is known about the mechanism of AA5 enzymes comes from studies on the archetype, FgrGalOx. AA5 enzymes oxidize their substrates through a ping-pong mechanism involving a corresponding reduction of oxygen to hydrogen peroxide mediated by a mononuclear copper center, which is complexed via a distinct, crosslinked tyrosyl-cysteine residue (see below) [1, 4, 18, 19, 40, 41, 42, 43]. The first half-reaction results in a two-electron oxidation of the substrate and corresponding reduction of the Cu[II]-tyrosyl radical to a Cu[I]-tyrosine (phenol). The second half-reaction regenerates the oxidation state of the active-site through reduction of molecular oxygen to hydrogen peroxide. Detailed kinetic studies, including kinetic isotope effects, suggest that each half reaction consists of three steps: proton transfer (PT), hydrogen atom transfer (HAT), and electron transfer (ET) [42, 44]. Due to its fundamental uniqueness, the mechanism of AA5 CROs has received significant theoretical treatment and the synthesis of many chemical mimetics has been attempted [45, 46].
Practically, AA5 enzymes are conveniently assayed by measuring hydrogen peroxide (co-product) generatation, e.g. in coupled reactions with horseradish peroxidase and a chromogenic substrate. In preparative reactions, catalase is typically added to prevent accumulation of hydrogen peroxide. AA5 enzymes are prone to inactivation by one-electron reduction to a Cu[I]-tyrosyl radical. The resulting off-cycle species can be rescued by oxidation by peroxidases or transition metal ions (ferricyanide, Mg(III), etc.), the inclusion of which in reactions is required to obtain maximal activity and substrate conversion [39, 47, 48, 49, 50, 51].
Catalytic Residues
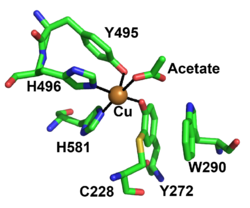
The redox-active center of AA5 oxidases comprises a copper ion that is stabilized by two tyrosines and two histidines (in FgrGalOx, these are Tyr272, Tyr495, His496, His581), resulting in a distorted square pyramidal geometry [4, 18, 19, 40]. Based on the copper coordination environment, AA5 proteins are type 2 “non-blue” copper category due to the nitrogen and oxygen ligands [52]. The unique feature of AA5 enzymes is the covalently linked equatorial tyrosine with an adjacent cysteine by a thioether bond (Tyr 272 and Cys 228 in the archetypal FgrGalOx) [52, 53]. The thioether linkage forms spontaneously in the presence of copper and has been shown to stabilize the radical though delocalization that forms on the equatorial tyrosine during catalysis [54].
Another important feature of AA5 enzymes is a secondary shell amino acid that is located on top of the tyrosine-cysteine cofactor. It has been speculated to be critical in determining the substrate specificity, radical stability and redox activity by hydrogen bonding, delocalization and/or by protecting the thioether bond from solvent [1, 3, 4, 55]. This residue in AA5_1, based on sequence alignments, has been conserved as a histidine [1], while characterized AA5_2 enzymes have an aromatic residue at this position: a tryptophan in galactose oxidases (W290 in FgrGalOx) [6, 55], a phenylalanine in the Colletotrichum aliphatic alcohol oxidases [7], whereas a tyrosine is present in the raffinose oxidases [5, 6] and aryl alcohol oxidase from Colletotrichum graminicola [9]. Furthermore, an AA5 enzyme from Streptomyces lividans with activity on glycolaldehyde possesses a tryptophan as the stacking secondary shell residue whose indole ring is oriented differently compared to FgrGalOx, which may affect the substrate specificity [17, 56].
In AA5_2 the Tyr-Cys cofactor exhibits an unusually low reduction potential (+275 mV) [57, 58, 59] compared to unmodified tyrosine in solution (> +800 mV) or in other enzymatic systems [60]. Several factors could contribute to this phenomenon by increasing the stability of the protein free radical including π-stacking with aromatic residues and the electron donating effect of the thioether linkage [4, 55, 61]. In contrast, AA5_1 have a reduction potential around +640 mV [18] which could explain the different oxidizing power of these two subfamilies [1, 59]. One reason for the higher reduction potential of glyoxal oxidases could be subtitution of the secondary shell amino acid Trp in AA5_2 with a His in AA5_1 [1, 59]. In the archetypal AA5_2 member, FgrGalOx, the Trp290His substitution increased the reduction potential of the resulting enzyme from +400 mV to +730 mV [62]; however, it also decreased the catalytic efficiency by 1000-fold [41] and affected the stability of the [Cu2+ Tyr·] metallo-radical complex at neutral pH [63]. CgrAlcOx and CgrAAO have been speculated to have a lower reduction potential than FgrGalOx due to their secondary shell amino acid substitutions (Phe in CgrAlcOx and Tyr in CgrAAO) [7, 9].
Three-dimensional Structures
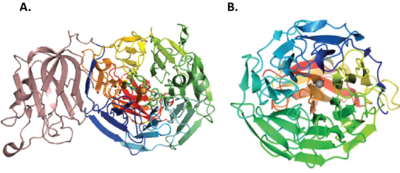
AA5s share a seven-bladed β-propeller fold [7, 9, 52] for the catalytic domain containing the active site. The archetypal FgrGalOx contains three domains: domain 1 has a “β sandwich” structure identified as a carbohydrate binding module (CBM32) with affinity for galactose, domain 2 is the catalytic domain and domain 3 is the smallest, which forms a hydrogen bonding network to stabilize domain 2 [52]. Other characterized AA5_2 enzymes from Fusarium species contain CBM32 [6, 23, 25, 64], even though some do not display canonical galactose oxidase activity (ex. FgrAAO and FoxAAO) [6, 10]. In contrast, CgrAlcOx, CglAlcOx and ChiAlcOx do not possess any CBM [7, 8], while CgrAAO and CgrRafOx have a PAN domain present instead [5, 9]. PorAlcOx contained a WSC domain that was able to bind xylans and fungal chitin/β-1,3-glucan, implicating the domain's involvement in enzyme anchoring on the plant surface [8]. In addition, the fusion of a galactose oxidase with a CBM29 has shown to increase the catalytic efficiency of the construct on galactose-containing hemicelluloses compared to WT [65].
Family Firsts
- First AA5_1 enzyme discovered
- The glyoxal oxidase from Phanerochaete chrysosporium discovered in 1987 [14].
- First AA5_2 enzyme discovered
- The archetypal galactose-6 oxidase from Fusarium graminearum (FgrGalOx) discovered in 1959 [12].
- Copper requirement confirmed
- While this first report already established FgrGalOx as a metalloenzyme; its copper requirement was later confirmed [66].
- First 3-D structure
- The first crystallography structure of AA5 was of the archetypal FgrGalOx in 1991 [53].
References
- Kersten P and Cullen D. (2014). Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet Biol. 2014;72:124-130. DOI:10.1016/j.fgb.2014.05.011 |
- Levasseur A, Drula E, Lombard V, Coutinho PM, and Henrissat B. (2013). Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6(1):41. DOI:10.1186/1754-6834-6-41 |
- Daou M and Faulds CB. (2017). Glyoxal oxidases: their nature and properties. World J Microbiol Biotechnol. 2017;33(5):87. DOI:10.1007/s11274-017-2254-1 |
- Whittaker JW (2003). Free radical catalysis by galactose oxidase. Chem Rev. 2003;103(6):2347-63. DOI:10.1021/cr020425z |
- Andberg M, Mollerup F, Parikka K, Koutaniemi S, Boer H, Juvonen M, Master E, Tenkanen M, and Kruus K. (2017). A Novel Colletotrichum graminicola Raffinose Oxidase in the AA5 Family. Appl Environ Microbiol. 2017;83(20). DOI:10.1128/AEM.01383-17 |
-
pmid=
- Yin DT, Urresti S, Lafond M, Johnston EM, Derikvand F, Ciano L, Berrin JG, Henrissat B, Walton PH, Davies GJ, and Brumer H. (2015). Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family. Nat Commun. 2015;6:10197. DOI:10.1038/ncomms10197 |
- Oide S, Tanaka Y, Watanabe A, and Inui M. (2019). Carbohydrate-binding property of a cell wall integrity and stress response component (WSC) domain of an alcohol oxidase from the rice blast pathogen Pyricularia oryzae. Enzyme Microb Technol. 2019;125:13-20. DOI:10.1016/j.enzmictec.2019.02.009 |
-
Mathieu, Y., Offen, W. A., Forget, S. M., Ciano, L., Viborg, A. H., Blagova, E., Henrissat, B., Walton, P.H, Davies, G.J, and Brumer, H. (2020). Discovery of a fungal copper radical oxidase with high catalytic efficiency toward 5-hydroxymethylfurfural and benzyl alcohols for bioprocessing. ACS Catalysis, 10, 3042-3058. DOI:10.1021/acscatal.9b04727
- Cleveland M, Lafond M, Xia FR, Chung R, Mulyk P, Hein JE, and Brumer H. (2021). Two Fusarium copper radical oxidases with high activity on aryl alcohols. Biotechnol Biofuels. 2021;14(1):138. DOI:10.1186/s13068-021-01984-0 |
-
Ögel, Z. B.; Brayford, D.; McPherson, M. J., (1994). Cellulose-triggered sporulation in the galactose oxidase-producing fungus Cladobotryum (Dactylium) dendroides NRRL 2903 and its re-identification as a species of Fusarium. Mycol. Res., 98, 474-480. DOI:10.1016/S0953-7562(09)81207-0
- COOPER JA, SMITH W, BACILA M, and MEDINA H. (1959). Galactose oxidase from Polyporus circinatus, Fr. J Biol Chem. 1959;234(3):445-8. | Google Books | Open Library
- AVIGAD G, AMARAL D, ASENSIO C, and HORECKER BL. (1962). The D-galactose oxidase of Polyporus circinatus. J Biol Chem. 1962;237:2736-43. | Google Books | Open Library
- Kersten PJ and Kirk TK. (1987). Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J Bacteriol. 1987;169(5):2195-201. DOI:10.1128/jb.169.5.2195-2201.1987 |
- Šola K, Gilchrist EJ, Ropartz D, Wang L, Feussner I, Mansfield SD, Ralet MC, and Haughn GW. (2019). RUBY, a Putative Galactose Oxidase, Influences Pectin Properties and Promotes Cell-To-Cell Adhesion in the Seed Coat Epidermis of Arabidopsis. Plant Cell. 2019;31(4):809-831. DOI:10.1105/tpc.18.00954 |
- Šola K, Dean GH, Li Y, Lohmann J, Movahedan M, Gilchrist EJ, Adams KL, and Haughn GW. (2021). Expression Patterns and Functional Characterization of Arabidopsis Galactose Oxidase-Like Genes Suggest Specialized Roles for Galactose Oxidases in Plants. Plant Cell Physiol. 2021;62(12):1927-1943. DOI:10.1093/pcp/pcab073 |
- Chaplin AK, Petrus ML, Mangiameli G, Hough MA, Svistunenko DA, Nicholls P, Claessen D, Vijgenboom E, and Worrall JA. (2015). GlxA is a new structural member of the radical copper oxidase family and is required for glycan deposition at hyphal tips and morphogenesis of Streptomyces lividans. Biochem J. 2015;469(3):433-44. DOI:10.1042/BJ20150190 |
- Whittaker MM, Kersten PJ, Nakamura N, Sanders-Loehr J, Schweizer ES, and Whittaker JW. (1996). Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J Biol Chem. 1996;271(2):681-7. DOI:10.1074/jbc.271.2.681 |
- Whittaker MM, Kersten PJ, Cullen D, and Whittaker JW. (1999). Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J Biol Chem. 1999;274(51):36226-32. DOI:10.1074/jbc.274.51.36226 |
- Leuthner B, Aichinger C, Oehmen E, Koopmann E, Müller O, Müller P, Kahmann R, Bölker M, and Schreier PH. (2005). A H2O2-producing glyoxal oxidase is required for filamentous growth and pathogenicity in Ustilago maydis. Mol Genet Genomics. 2005;272(6):639-50. DOI:10.1007/s00438-004-1085-6 |
- Daou M, Piumi F, Cullen D, Record E, and Faulds CB. (2016). Heterologous Production and Characterization of Two Glyoxal Oxidases from Pycnoporus cinnabarinus. Appl Environ Microbiol. 2016;82(16):4867-75. DOI:10.1128/AEM.00304-16 |
-
Parikka, K.; Master, E.; Tenkanen, M., (2015) Oxidation with galactose oxidase: Multifunctional enzymatic catalysis. J. Mol. Catal. B: Enzym. 120, 47-59. https://doi.org/10.1016/j.molcatb.2015.06.006
- Paukner R, Staudigl P, Choosri W, Haltrich D, and Leitner C. (2015). Expression, purification, and characterization of galactose oxidase of Fusarium sambucinum in E. coli. Protein Expr Purif. 2015;108:73-79. DOI:10.1016/j.pep.2014.12.010 |
- Parikka K, Leppänen AS, Pitkänen L, Reunanen M, Willför S, and Tenkanen M. (2010). Oxidation of polysaccharides by galactose oxidase. J Agric Food Chem. 2010;58(1):262-71. DOI:10.1021/jf902930t |
- Paukner R, Staudigl P, Choosri W, Sygmund C, Halada P, Haltrich D, and Leitner C. (2014). Galactose oxidase from Fusarium oxysporum--expression in E. coli and P. pastoris and biochemical characterization. PLoS One. 2014;9(6):e100116. DOI:10.1371/journal.pone.0100116 |
- Alberton D, Silva de Oliveira L, Peralta RM, and Barbosa-Tessmann IP. (2007). Production, purification, and characterization of a novel galactose oxidase from Fusarium acuminatum. J Basic Microbiol. 2007;47(3):203-12. DOI:10.1002/jobm.200610290 |
- Minasian SG, Whittaker MM, and Whittaker JW. (2004). Stereoselective hydrogen abstraction by galactose oxidase. Biochemistry. 2004;43(43):13683-93. DOI:10.1021/bi048554s |
-
Siebum, A., van Wijk, A., Schoevaart, R. & Kieboom, T. (2006) Galactose oxidase and alcohol oxidase: scope and limitations for the enzymatic synthesis of aldehydes. J. Mol. Catal. B, 41, 141–145. DOI:10.1016/j.molcatb.2006.04.003
-
Yalpani, M.; Hall, L. D., (1982) Some chemical and analytical aspects of polysaccharide modifications. II. A high-yielding, specific method for the chemical derivatization of galactose-containing polysaccharides: Oxidation with galactose oxidase followed by reductive amination. J. Polym. Sci., Polym. Chem. Ed. 20, 3399-3420. https://doi.org/10.1002/pol.1982.170201213
- Kelleher FM and Bhavanandan VP. (1986). Preparation and characterization of beta-D-fructofuranosyl O-(alpha-D-galactopyranosyl uronic acid)-(1----6)-O-alpha-D-glucopyranoside and O-(alpha-D-galactopyranosyl uronic acid)-(1----6)-D-glucose. Carbohydr Res. 1986;155:89-97. DOI:10.1016/s0008-6215(00)90135-6 |
-
Schoevaart, R.; Kieboom, T., (2004) Application of Galactose Oxidase in Chemoenzymatic One-Pot Cascade Reactions Without Intermediate Recovery Steps. Top. Catal. 27, 3-9. https://doi.org/10.1023/B:TOCA.0000013536.27551.13
- Leppänen AS, Xu C, Parikka K, Eklund P, Sjöholm R, Brumer H, Tenkanen M, and Willför S. (2014). Targeted allylation and propargylation of galactose-containing polysaccharides in water. Carbohydr Polym. 2014;100:46-54. DOI:10.1016/j.carbpol.2012.11.053 |
- Xu C, Spadiut O, Araújo AC, Nakhai A, and Brumer H. (2012). Chemo-enzymatic assembly of clickable cellulose surfaces via multivalent polysaccharides. ChemSusChem. 2012;5(4):661-5. DOI:10.1002/cssc.201100522 |
- Parikka K, Leppänen AS, Xu C, Pitkänen L, Eronen P, Osterberg M, Brumer H, Willför S, and Tenkanen M. (2012). Functional and anionic cellulose-interacting polymers by selective chemo-enzymatic carboxylation of galactose-containing polysaccharides. Biomacromolecules. 2012;13(8):2418-28. DOI:10.1021/bm300679a |
-
Mikkonen, K. S.; Parikka, K.; Suuronen, J.-P.; Ghafar, A.; Serimaa, R.; Tenkanen, M., (2014) Enzymatic oxidation as a potential new route to produce polysaccharide aerogels. RSC Advances. 4, 11884-11892. https://doi.org/10.1039/C3RA47440B
- Derikvand F, Yin DT, Barrett R, and Brumer H. (2016). Cellulose-Based Biosensors for Esterase Detection. Anal Chem. 2016;88(6):2989-93. DOI:10.1021/acs.analchem.5b04661 |
- Ribeaucourt D, Saker S, Navarro D, Bissaro B, Drula E, Correia LO, Haon M, Grisel S, Lapalu N, Henrissat B, O'Connell RJ, Lambert F, Lafond M, and Berrin JG. (2021). Identification of Copper-Containing Oxidoreductases in the Secretomes of Three Colletotrichum Species with a Focus on Copper Radical Oxidases for the Biocatalytic Production of Fatty Aldehydes. Appl Environ Microbiol. 2021;87(24):e0152621. DOI:10.1128/AEM.01526-21 |
- Ribeaucourt D, Bissaro B, Lambert F, Lafond M, and Berrin JG. (2022). Biocatalytic oxidation of fatty alcohols into aldehydes for the flavors and fragrances industry. Biotechnol Adv. 2022;56:107787. DOI:10.1016/j.biotechadv.2021.107787 |
- Roncal T, Muñoz C, Lorenzo L, Maestro B, and Díaz de Guereñu Mdel M. (2012). Two-step oxidation of glycerol to glyceric acid catalyzed by the Phanerochaete chrysosporium glyoxal oxidase. Enzyme Microb Technol. 2012;50(2):143-50. DOI:10.1016/j.enzmictec.2011.11.007 |
- Whittaker JW (2005). The radical chemistry of galactose oxidase. Arch Biochem Biophys. 2005;433(1):227-39. DOI:10.1016/j.abb.2004.08.034 |
- Baron AJ, Stevens C, Wilmot C, Seneviratne KD, Blakeley V, Dooley DM, Phillips SE, Knowles PF, and McPherson MJ. (1994). Structure and mechanism of galactose oxidase. The free radical site. J Biol Chem. 1994;269(40):25095-105. | Google Books | Open Library
- Humphreys KJ, Mirica LM, Wang Y, and Klinman JP. (2009). Galactose oxidase as a model for reactivity at a copper superoxide center. J Am Chem Soc. 2009;131(13):4657-63. DOI:10.1021/ja807963e |
- Whittaker MM and Whittaker JW. (1993). Ligand interactions with galactose oxidase: mechanistic insights. Biophys J. 1993;64(3):762-72. DOI:10.1016/S0006-3495(93)81437-1 |
- Whittaker MM, Ballou DP, and Whittaker JW. (1998). Kinetic isotope effects as probes of the mechanism of galactose oxidase. Biochemistry. 1998;37(23):8426-36. DOI:10.1021/bi980328t |
- Wang Y, DuBois JL, Hedman B, Hodgson KO, and Stack TD. (1998). Catalytic galactose oxidase models: biomimetic Cu(II)-phenoxyl-radical reactivity. Science. 1998;279(5350):537-40. DOI:10.1126/science.279.5350.537 |
-
Himo, F.; Eriksson, L. A.; Maseras, F.; Siegbahn, P. E. M., (2000). Catalytic Mechanism of Galactose Oxidase: A Theoretical Study. J. Am. Chem. Soc. 122, 8031-8036. https://doi.org/10.1021/ja994527r
- Cleveland L, Coffman RE, Coon P, and Davis L. (1975). An investigation of the role of the copper in galactose oxidase. Biochemistry. 1975;14(6):1108-15. DOI:10.1021/bi00677a003 |
- Hamilton GA, Dyrkacz GR, and Libby RD. (1976). The involvement of superoxide and trivalent copper in the galactose oxidase reaction. Adv Exp Med Biol. 1976;74:489-504. DOI:10.1007/978-1-4684-3270-1_42 |
-
Toftgaard Pedersen, A.; Birmingham, W. R.; Rehn, G.; Charnock, S. J.; Turner, N. J.; Woodley, J. M., (2015) Process Requirements of Galactose Oxidase Catalyzed Oxidation of Alcohols. Org. Process Res. Dev. 19, 1580-1589. https://doi.org/10.1021/acs.oprd.5b00278
- Forget SM, Xia FR, Hein JE, and Brumer H. (2020). Determination of biocatalytic parameters of a copper radical oxidase using real-time reaction progress monitoring. Org Biomol Chem. 2020;18(11):2076-2084. DOI:10.1039/c9ob02757b |
- Johnson HC, Zhang S, Fryszkowska A, Ruccolo S, Robaire SA, Klapars A, Patel NR, Whittaker AM, Huffman MA, and Strotman NA. (2021). Biocatalytic oxidation of alcohols using galactose oxidase and a manganese(III) activator for the synthesis of islatravir. Org Biomol Chem. 2021;19(7):1620-1625. DOI:10.1039/d0ob02395g |
- Ito N, Phillips SE, Yadav KD, and Knowles PF. (1994). Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol. 1994;238(5):794-814. DOI:10.1006/jmbi.1994.1335 |
- Ito N, Phillips SE, Stevens C, Ogel ZB, McPherson MJ, Keen JN, Yadav KD, and Knowles PF. (1991). Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature. 1991;350(6313):87-90. DOI:10.1038/350087a0 |
- Rogers MS, Hurtado-Guerrero R, Firbank SJ, Halcrow MA, Dooley DM, Phillips SE, Knowles PF, and McPherson MJ. (2008). Cross-link formation of the cysteine 228-tyrosine 272 catalytic cofactor of galactose oxidase does not require dioxygen. Biochemistry. 2008;47(39):10428-39. DOI:10.1021/bi8010835 |
- Rogers MS, Tyler EM, Akyumani N, Kurtis CR, Spooner RK, Deacon SE, Tamber S, Firbank SJ, Mahmoud K, Knowles PF, Phillips SE, McPherson MJ, and Dooley DM. (2007). The stacking tryptophan of galactose oxidase: a second-coordination sphere residue that has profound effects on tyrosyl radical behavior and enzyme catalysis. Biochemistry. 2007;46(15):4606-18. DOI:10.1021/bi062139d |
- Chaplin AK, Svistunenko DA, Hough MA, Wilson MT, Vijgenboom E, and Worrall JA. (2017). Active-site maturation and activity of the copper-radical oxidase GlxA are governed by a tryptophan residue. Biochem J. 2017;474(5):809-825. DOI:10.1042/BCJ20160968 |
- Cowley RE, Cirera J, Qayyum MF, Rokhsana D, Hedman B, Hodgson KO, Dooley DM, and Solomon EI. (2016). Structure of the Reduced Copper Active Site in Preprocessed Galactose Oxidase: Ligand Tuning for One-Electron O(2) Activation in Cofactor Biogenesis. J Am Chem Soc. 2016;138(40):13219-13229. DOI:10.1021/jacs.6b05792 |
- Thomas F, Gellon G, Gautier-Luneau I, Saint-Aman E, and Pierre JL. (2002). A structural and functional model of galactose oxidase: control of the one-electron oxidized active form through two differentiated phenolic arms in a tripodal ligand. Angew Chem Int Ed Engl. 2002;41(16):3047-50. DOI:10.1002/1521-3773(20020816)41:16<3047::AID-ANIE3047>3.0.CO;2-W |
- Wright C and Sykes AG. (2001). Interconversion of Cu(I) and Cu(II) forms of galactose oxidase: comparison of reduction potentials. J Inorg Biochem. 2001;85(4):237-43. DOI:10.1016/s0162-0134(01)00214-8 |
-
Itoh, S.; Taki, M.; Fukuzumi, S., (2000). Active site models for galactose oxidase and related enzymes. Coord. Chem. Rev. 198, 3-20. https://doi.org/10.1016/S0010-8545(99)00209-X
-
Jazdzewski, B. A.; Tolman, W. B., (2000). Understanding the copper–phenoxyl radical array in galactose oxidase: contributions from synthetic modeling studies. Coord. Chem. Rev. 200-202, 633-685. https://doi.org/10.1016/S0010-8545(00)00342-8
-
Saysell, C. G.; Barna, T.; Borman, C. D.; Baron, A. J.; McPherson, M. J.; Sykes, A. G., P(1997). Properties of the Trp290His variant of Fusarium NRRL 2903 galactose oxidase: interactions of the GOasesemi state with different buffers, its redox activity and ability to bind azide. J. Biol. Inorg. Chem. 2, 702-709. https://doi.org/10.1007/s007750050186
-
Rogers, M. S.; Knowles, P. F.; Baron, A. J.; McPherson, M. J.; Dooley, D. M., (1998). Characterization of the active site of galactose oxidase and its active site mutational variants Y495F/H/K and W290H by circular dichroism spectroscopy. Inorg. Chim. Acta. 275-276, 175-181. https://doi.org/10.1016/S0020-1693(97)06142-2
- Faria CB, de Castro FF, Martim DB, Abe CAL, Prates KV, de Oliveira MAS, and Barbosa-Tessmann IP. (2019). Production of Galactose Oxidase Inside the Fusarium fujikuroi Species Complex and Recombinant Expression and Characterization of the Galactose Oxidase GaoA Protein from Fusarium subglutinans. Mol Biotechnol. 2019;61(9):633-649. DOI:10.1007/s12033-019-00190-6 |
- Mollerup F and Master E. (2016). Influence of a family 29 carbohydrate binding module on the recombinant production of galactose oxidase in Pichia pastoris. Data Brief. 2016;6:176-83. DOI:10.1016/j.dib.2015.11.032 |
- AMARAL D, BERNSTEIN L, MORSE D, and HORECKER BL. (1963). Galactose oxidase of Polyporus circinatus: a copper enzyme. J Biol Chem. 1963;238:2281-4. | Google Books | Open Library
- Kersten PJ (1990). Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. Proc Natl Acad Sci U S A. 1990;87(8):2936-40. DOI:10.1073/pnas.87.8.2936 |