CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Carbohydrate Binding Module Family 22"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | * [[Author]]: | + | * [[Author]]: [[User:Harry Gilbert|Harry Gilbert]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Elizabeth Ficko-Blean|Elizabeth Ficko-Blean]] |
---- | ---- | ||
| Line 17: | Line 17: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | CBM22 is predominantly a bacterial family of protein modules that comprises around 150 amino acids. The primary target for CBM22 members is xylan, although they bind <100-fold weaker to soluble β1,4-glucans and β1,3-β1,4-mixed linked glucans <cite>Charnock2000,Wassenberg2000, Sainz-Polo2015</cite>. There is one report showing significant qualitative binding of a CBM22, derived from a ruminal protozoan GH10 xylanase, to different forms of crystalline cellulose <cite>Devillard2003</cite> . CBM22s from thermophilic and mesophilic organisms have a ''K''<sub>D</sub> for xylan of 2 µM and 100 µM, respectively. Binding studies of xylooligosaccharides indicated that these CBMs contain four <cite>Charnock2000,Wassenberg2000</cite> or five <cite>Sainz-Polo2015</cite> xylose binding sites. Isothermal titration calorimetry showed that ligand binding was enthalpically driven with changes in entropy having a negative impact on affinity. Coverage of polysaccharide ligands at saturation indicated that CBM22 members are endo binding. CBM22 is thus classified as a type B CBM22. | + | CBM22 is predominantly a bacterial family of protein modules that comprises around 150 amino acids. The primary target for CBM22 members is xylan, although they bind <100-fold weaker to soluble β1,4-glucans and β1,3-β1,4-mixed linked glucans <cite>Charnock2000,Wassenberg2000, Sainz-Polo2015</cite>. There is one report showing significant qualitative binding of a CBM22, derived from a ruminal protozoan GH10 xylanase, to different forms of crystalline cellulose <cite>Devillard2003</cite> . CBM22s from thermophilic and mesophilic organisms have a ''K''<sub>D</sub> for xylan of 2 µM and 100 µM, respectively. Binding studies of xylooligosaccharides indicated that these CBMs contain four <cite>Charnock2000,Wassenberg2000</cite> or five <cite>Sainz-Polo2015</cite> xylose binding sites. Isothermal titration calorimetry showed that ligand binding was enthalpically driven with changes in entropy having a negative impact on affinity. Coverage of polysaccharide ligands at saturation indicated that CBM22 members are endo binding. CBM22 is thus classified as a [[Carbohydrate-binding_modules#Types|type B]] CBM22. |
== Structural Features == | == Structural Features == | ||
[[File:cbm22.png|thumb|300px|right|'''Figure 1.''' The fold and ligand binding cleft of CBM22-2 from the ''Paenibacillus barcinonensis'' xylanase Xyn10C (see [{{PDBlink}}4XUR PDB ID 4XUR]) highlighting the topography of the binding site and the location of key residues in ligand recognition.]] | [[File:cbm22.png|thumb|300px|right|'''Figure 1.''' The fold and ligand binding cleft of CBM22-2 from the ''Paenibacillus barcinonensis'' xylanase Xyn10C (see [{{PDBlink}}4XUR PDB ID 4XUR]) highlighting the topography of the binding site and the location of key residues in ligand recognition.]] | ||
| − | The structures of CBM22 members adopt a canonical β-sandwich fold in which the two β-sheets both contain five antiparallel β-strands <cite>Charnock2000, Sainz-Polo2015</cite>. One of the β-sheets presents a surface groove that runs along the whole of the concave face of the β-sandwich. This deep cleft is approximately 20 Å long and has a depth of 9 Å in the C-terminal CBM22 (Xyn10B-CBM22-2) in the ''Clostridium thermocellum'' xylanase Xyn10B. The dimensions of the cleft corresponds to the length of a xylotetraose moiety and are consistent with biochemical data showing that the ligand binding site interacts with four xylose binding units <cite>Charnock2000</cite>. The interactions between a CBM22 and its xylan ligand was revealed by the structure of CBM22-2 (Xyn10C-CBM22-2) from the ''Paenibacillus barcinonensis'' xylanase Xyn10C in complex with xylotetraose ('''Figure 1''') <cite>Sainz-Polo2015</cite>. The structure showed that the ligand was in its expected 3-fold helical conformation and that in the centre of the binding site two highly conserved aromatic residues made planar hydrophobic interactions with xylose sugars n and n+1 (i.e. adjacent xylose residues). These apolar interactions were augmented by a limited number of hydrogen bonds with four conserved polar amino acids <cite>Sainz-Polo2015</cite>. Mutagenesis data confirmed the functional importance of these conserved aromatic and polar residues <cite>Xie2001</cite>. Several O2 and O3 groups of the xylose residues are solvent exposed explaining why CBM22 members are able to bind to highly decorated xylans such as wheat and rye arabinoxylan. A significant difference between Xyn10C-CBM22-2 and Xyn10B-CBM22-2 is that in the ''Paenibacillus'' module an extended loop presents an additional tryptophan that stacks against xylose n+1, which is thus sandwiched between two aromatic residues. | + | The structures of CBM22 members adopt a canonical β-sandwich fold in which the two β-sheets both contain five antiparallel β-strands <cite>Charnock2000, Sainz-Polo2015</cite>. One of the β-sheets presents a surface groove that runs along the whole of the concave face of the β-sandwich. This deep cleft is approximately 20 Å long and has a depth of 9 Å in the C-terminal CBM22 (Xyn10B-CBM22-2) in the ''Clostridium thermocellum'' xylanase Xyn10B. The dimensions of the cleft corresponds to the length of a xylotetraose moiety and are consistent with biochemical data showing that the ligand binding site interacts with four xylose binding units <cite>Charnock2000</cite>. The interactions between a CBM22 module and its xylan ligand was revealed by the structure of CBM22-2 (Xyn10C-CBM22-2) from the ''Paenibacillus barcinonensis'' xylanase Xyn10C in complex with xylotetraose ('''Figure 1''') <cite>Sainz-Polo2015</cite>. The structure showed that the ligand was in its expected 3-fold helical conformation and that in the centre of the binding site two highly conserved aromatic residues made planar hydrophobic interactions with xylose sugars n and n+1 (i.e. adjacent xylose residues). These apolar interactions were augmented by a limited number of hydrogen bonds with four conserved polar amino acids <cite>Sainz-Polo2015</cite>. Mutagenesis data confirmed the functional importance of these conserved aromatic and polar residues <cite>Xie2001</cite>. Several O2 and O3 groups of the xylose residues are solvent exposed explaining why CBM22 members are able to bind to highly decorated xylans such as wheat and rye arabinoxylan. A significant difference between Xyn10C-CBM22-2 and Xyn10B-CBM22-2 is that in the ''Paenibacillus'' module an extended loop presents an additional tryptophan that stacks against xylose n+1, which is thus sandwiched between two aromatic residues. |
== Functionalities == | == Functionalities == | ||
| − | The vast majority of CBM22 members are located in GH10 xylanases. These enzymes often contain multiple copies of CBM22s, and frequently additional CBMs, particularly members of [[CBM9]] <cite>Wassenberg2000,Sainz-Polo2015</cite>. Prior to the discovery of their carbohydrate binding function, CBM22s were defined as X6 modules and were shown to confer substantial an increase in thermostability, which was initially thought to be the major function of these modules <cite>Fontes1995,Winterhalter1995,Lee1993</cite>, and is still thought to be of functional significance <cite>Meng2015</cite>. Despite multiple copies of CBM22s in many xylanases there have been no reports of increased affinity through avidity effects <cite>Charnock2000,Wassenberg2000, Sainz-Polo2015</cite>. In general the multiple copies of CBM22s in a specific enzyme display similar ligand binding properties <cite>Sainz-Polo2015</cite>, although the role, if any, of some members of this family is unknown <cite>Najmudin2010</cite>. Cell labelling studies showed that Xyn10B-CBM22-2 displayed weak binding to plant cell wall sections, possibly reflecting stacking of aromatic residues to the α and β face of the same xylose, which may prevent access to xylans embedded in the complex milieu of the plant cell wall <cite>McCartney2006</cite>. It was suggested that CBM22s primary targets are soluble xylans <cite>McCartney2006</cite>. | + | The vast majority of CBM22 members are located in GH10 xylanases. These enzymes often contain multiple copies of CBM22s, and frequently additional CBMs, particularly members of [[CBM9]] <cite>Wassenberg2000,Sainz-Polo2015</cite>. Prior to the discovery of their carbohydrate binding function, CBM22s were defined as X6 modules and were shown to confer substantial an increase in thermostability, which was initially thought to be the major function of these modules <cite>Fontes1995,Winterhalter1995,Lee1993</cite>, and is still thought to be of functional significance <cite>Meng2015</cite>. Despite multiple copies of CBM22s in many xylanases there have been no reports of increased affinity through avidity effects <cite>Charnock2000,Wassenberg2000, Sainz-Polo2015</cite>. In general the multiple copies of CBM22s in a specific enzyme display similar ligand binding properties <cite>Sainz-Polo2015</cite>, although the role, if any, of some members of this family is unknown <cite>Najmudin2010</cite>. Cell labelling studies showed that Xyn10B-CBM22-2 displayed weak binding to plant cell wall sections, possibly reflecting stacking of aromatic residues to the α and β face of the same xylose, which may prevent access to xylans embedded in the complex milieu of the plant cell wall <cite>McCartney2006</cite>. It was suggested that CBM22s primary targets are soluble xylans <cite>McCartney2006</cite>. Functional analysis of several multi-modular enzymes led to the proposal that CBM22 members can increase the activity of some xylanases <cite>Ali2005,Zhao2005</cite>. |
== Family Firsts == | == Family Firsts == | ||
Latest revision as of 14:14, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| http://www.cazy.org/CBM22.html |
Ligand specificities
CBM22 is predominantly a bacterial family of protein modules that comprises around 150 amino acids. The primary target for CBM22 members is xylan, although they bind <100-fold weaker to soluble β1,4-glucans and β1,3-β1,4-mixed linked glucans [1, 2, 3]. There is one report showing significant qualitative binding of a CBM22, derived from a ruminal protozoan GH10 xylanase, to different forms of crystalline cellulose [4] . CBM22s from thermophilic and mesophilic organisms have a KD for xylan of 2 µM and 100 µM, respectively. Binding studies of xylooligosaccharides indicated that these CBMs contain four [1, 2] or five [3] xylose binding sites. Isothermal titration calorimetry showed that ligand binding was enthalpically driven with changes in entropy having a negative impact on affinity. Coverage of polysaccharide ligands at saturation indicated that CBM22 members are endo binding. CBM22 is thus classified as a type B CBM22.
Structural Features
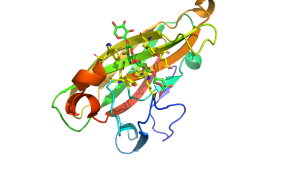
The structures of CBM22 members adopt a canonical β-sandwich fold in which the two β-sheets both contain five antiparallel β-strands [1, 3]. One of the β-sheets presents a surface groove that runs along the whole of the concave face of the β-sandwich. This deep cleft is approximately 20 Å long and has a depth of 9 Å in the C-terminal CBM22 (Xyn10B-CBM22-2) in the Clostridium thermocellum xylanase Xyn10B. The dimensions of the cleft corresponds to the length of a xylotetraose moiety and are consistent with biochemical data showing that the ligand binding site interacts with four xylose binding units [1]. The interactions between a CBM22 module and its xylan ligand was revealed by the structure of CBM22-2 (Xyn10C-CBM22-2) from the Paenibacillus barcinonensis xylanase Xyn10C in complex with xylotetraose (Figure 1) [3]. The structure showed that the ligand was in its expected 3-fold helical conformation and that in the centre of the binding site two highly conserved aromatic residues made planar hydrophobic interactions with xylose sugars n and n+1 (i.e. adjacent xylose residues). These apolar interactions were augmented by a limited number of hydrogen bonds with four conserved polar amino acids [3]. Mutagenesis data confirmed the functional importance of these conserved aromatic and polar residues [5]. Several O2 and O3 groups of the xylose residues are solvent exposed explaining why CBM22 members are able to bind to highly decorated xylans such as wheat and rye arabinoxylan. A significant difference between Xyn10C-CBM22-2 and Xyn10B-CBM22-2 is that in the Paenibacillus module an extended loop presents an additional tryptophan that stacks against xylose n+1, which is thus sandwiched between two aromatic residues.
Functionalities
The vast majority of CBM22 members are located in GH10 xylanases. These enzymes often contain multiple copies of CBM22s, and frequently additional CBMs, particularly members of CBM9 [2, 3]. Prior to the discovery of their carbohydrate binding function, CBM22s were defined as X6 modules and were shown to confer substantial an increase in thermostability, which was initially thought to be the major function of these modules [6, 7, 8], and is still thought to be of functional significance [9]. Despite multiple copies of CBM22s in many xylanases there have been no reports of increased affinity through avidity effects [1, 2, 3]. In general the multiple copies of CBM22s in a specific enzyme display similar ligand binding properties [3], although the role, if any, of some members of this family is unknown [10]. Cell labelling studies showed that Xyn10B-CBM22-2 displayed weak binding to plant cell wall sections, possibly reflecting stacking of aromatic residues to the α and β face of the same xylose, which may prevent access to xylans embedded in the complex milieu of the plant cell wall [11]. It was suggested that CBM22s primary targets are soluble xylans [11]. Functional analysis of several multi-modular enzymes led to the proposal that CBM22 members can increase the activity of some xylanases [12, 13].
Family Firsts
- First Identified
- Xyn10B-CBM22-2 [1], the second CBM22 in the Thermotoga maritima GH10 xylanase Xyl10A [2], and the CBM22 in the GH10 xylanase XynA from Caldibaccilus cellulovorans [14] were both shown to bind xylan in early 2000.
- First Structural Characterization
- The first structural characterization of a CBM22 was the crystal structure of Xyn10B-CBM22-2 [1]. The first structure of a CBM22 in complex with ligand was CBM22-2 from Paenibacillus barcinonensis Xyn10C [3].
References
- Charnock SJ, Bolam DN, Turkenburg JP, Gilbert HJ, Ferreira LM, Davies GJ, and Fontes CM. (2000). The X6 "thermostabilizing" domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry. 2000;39(17):5013-21. DOI:10.1021/bi992821q |
- Meissner K, Wassenberg D, and Liebl W. (2000). The thermostabilizing domain of the modular xylanase XynA of Thermotoga maritima represents a novel type of binding domain with affinity for soluble xylan and mixed-linkage beta-1,3/beta-1, 4-glucan. Mol Microbiol. 2000;36(4):898-912. DOI:10.1046/j.1365-2958.2000.01909.x |
- Sainz-Polo MA, González B, Menéndez M, Pastor FI, and Sanz-Aparicio J. (2015). Exploring Multimodularity in Plant Cell Wall Deconstruction: STRUCTURAL AND FUNCTIONAL ANALYSIS OF Xyn10C CONTAINING THE CBM22-1-CBM22-2 TANDEM. J Biol Chem. 2015;290(28):17116-30. DOI:10.1074/jbc.M115.659300 |
- Devillard E, Bera-Maillet C, Flint HJ, Scott KP, Newbold CJ, Wallace RJ, Jouany JP, and Forano E. (2003). Characterization of XYN10B, a modular xylanase from the ruminal protozoan Polyplastron multivesiculatum, with a family 22 carbohydrate-binding module that binds to cellulose. Biochem J. 2003;373(Pt 2):495-503. DOI:10.1042/BJ20021784 |
- Xie H, Gilbert HJ, Charnock SJ, Davies GJ, Williamson MP, Simpson PJ, Raghothama S, Fontes CM, Dias FM, Ferreira LM, and Bolam DN. (2001). Clostridium thermocellum Xyn10B carbohydrate-binding module 22-2: the role of conserved amino acids in ligand binding. Biochemistry. 2001;40(31):9167-76. DOI:10.1021/bi0106742 |
- Fontes CM, Hazlewood GP, Morag E, Hall J, Hirst BH, and Gilbert HJ. (1995). Evidence for a general role for non-catalytic thermostabilizing domains in xylanases from thermophilic bacteria. Biochem J. 1995;307 ( Pt 1)(Pt 1):151-8. DOI:10.1042/bj3070151 |
- Winterhalter C, Heinrich P, Candussio A, Wich G, and Liebl W. (1995). Identification of a novel cellulose-binding domain within the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol Microbiol. 1995;15(3):431-44. DOI:10.1111/j.1365-2958.1995.tb02257.x |
- Lee YE, Lowe SE, Henrissat B, and Zeikus JG. (1993). Characterization of the active site and thermostability regions of endoxylanase from Thermoanaerobacterium saccharolyticum B6A-RI. J Bacteriol. 1993;175(18):5890-8. DOI:10.1128/jb.175.18.5890-5898.1993 |
- Meng DD, Ying Y, Chen XH, Lu M, Ning K, Wang LS, and Li FL. (2015). Distinct roles for carbohydrate-binding modules of glycoside hydrolase 10 (GH10) and GH11 xylanases from Caldicellulosiruptor sp. strain F32 in thermostability and catalytic efficiency. Appl Environ Microbiol. 2015;81(6):2006-14. DOI:10.1128/AEM.03677-14 |
- Najmudin S, Pinheiro BA, Prates JA, Gilbert HJ, Romão MJ, and Fontes CM. (2010). Putting an N-terminal end to the Clostridium thermocellum xylanase Xyn10B story: crystal structure of the CBM22-1-GH10 modules complexed with xylohexaose. J Struct Biol. 2010;172(3):353-62. DOI:10.1016/j.jsb.2010.07.009 |
- McCartney L, Blake AW, Flint J, Bolam DN, Boraston AB, Gilbert HJ, and Knox JP. (2006). Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc Natl Acad Sci U S A. 2006;103(12):4765-70. DOI:10.1073/pnas.0508887103 |
- Ali E, Zhao G, Sakka M, Kimura T, Ohmiya K, and Sakka K. (2005). Functions of family-22 carbohydrate-binding module in Clostridium thermocellum Xyn10C. Biosci Biotechnol Biochem. 2005;69(1):160-5. DOI:10.1271/bbb.69.160 |
- Zhao G, Ali E, Araki R, Sakka M, Kimura T, and Sakka K. (2005). Function of the family-9 and family-22 carbohydrate-binding modules in a modular beta-1,3-1,4-glucanase/xylanase derived from Clostridium stercorarium Xyn10B. Biosci Biotechnol Biochem. 2005;69(8):1562-7. DOI:10.1271/bbb.69.1562 |
- Sunna A, Gibbs MD, and Bergquist PL. (2000). The thermostabilizing domain, XynA, of Caldibacillus cellulovorans xylanase is a xylan binding domain. Biochem J. 2000;346 Pt 3(Pt 3):583-6. | Google Books | Open Library