CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Carbohydrate Binding Module Family 28
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Shinya Fushinobu^^^
- Responsible Curator: ^^^Shinya Fushinobu^^^
| CAZy DB link | |
| http://www.cazy.org/CBM28.html |
Ligand specificities
This module was firstly classified in 2002 by a characterization study of the C-terminal module in GH5 Cel5A from Bacillus sp. 1139 (BspCBM28) [1]. Currently known CBM28s are solely from bacterial origins, mostly attached to cellulosomal endoglucanases in tandem with CBM17. They bind non-crystalline (or amorphous) part of cellulose, cellooligosaccharides, or β-1,3-1,4-glucans [1, 2].
Structural Features
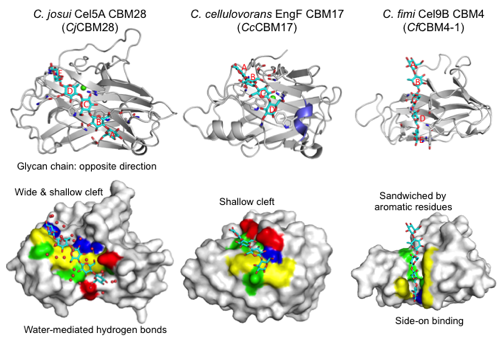
- Fold: CBM28s have a β-sandwich fold (approximately 200 amino acids) that is similar to CBM17 and CBM4 (Figure 1). The module has a straight cleft that binds a cellulose glycan chain at the center of the β-sandwich fold. A Ca2+ ion is bound on the back side of molecule. The Ca2+ ion is not involved in ligand binding but appears to play a structural stabilization role.
- Type: CBM28s are typical endo-type Type B CBMs that accommodate a single glycan chain because both ends of the cleft are open.
- Features of ligand binding: Figure 1 shows the cellopentaose complex structure (3aci) of CBM28 in GH5 Cel5A from Clostridium josui (CjCBM28). The long binding cleft runs at the center of the molecule. Therefore, the binding pocket location of CBM28 is on a face (side) of the β-sandwich fold, not on an apex/edge (CBM Fold). The shallow cleft of CBM28 binds one side of the cellooligosaccharides (face-on) in contrast with the case of CBM4 (side-on). There are at at least five subsites (A-E from the non-reducing end to the reducing end) in CjCBM28. Interestingly, the direction of the cellooligosaccharides bound to CBM28 is opposite to those in CBM17 and CBM4. Subsites B, C, and E form stacking interactions with aromatic residues (W78, W129, and F128 in CjCBM28). The flanking hydroxyl groups are extensively recognized by direct or water-mediated hydrogen bonds. Therefore, CBM28 has a relatively wide cleft.
Functionalities
- Functional role of CBM: CBM28s are thought to target catalytic modules (endoglucanases) to non-crystalline region of cellulose. Deletion mutants of CBM28 in Cel5A from Bacillus sp. 1139 showed significantly decreased amounts of the soluble products from amorphous cellulose [2].
- Most Common Associated Modules: Endo-β-1,4-glucanases of GH5. A survey using the GH-CBM tool shows that CBM28s are not associated with other GH family domains.
- Binding affinities: As a typical endo-type (Type B) CBM, CBM28s show the Ka values of 0.7-5.2 ×104 M-1 for the binding of cellooligosaccharides (cellotetraose to cellohexaose), and it is enthalpically driven [1, 6]. BspCBM28 binds amorphous (regenerated) cellulose with high-affinity (Ka = 9.9 ×105 M-1) and low-affinity (Ka = 2.1 ×104 M-1) cites [2].
- Novel Applications: BspCBM28 was used as the most amorphous-specific CBM in a study of photoactivated localization microscopy (PALM) [7].
Family Firsts
- First Identified
The first identified member is the C-terminal module in Cel5A from Bacillus sp. 1139 (BspCBM28) [1]
- First Structural Characterization
Partial assignment of the NMR data of a CBM28 in Cel5I from Clostridium cellulolyticum was reported in 2002 [8] but its three-dimensional structure is not reported yet. The first crystal structure was reported in 2004 for BspCBM28 in a ligand-free form (1uww) [9]. The first ligand complex structures were reported in 2010 for CBM28 in Cel5A from C. josui (CjCBM28) (3acf 3acg 3ach 3aci) [3].
References
- Boraston AB, Ghaffari M, Warren RA, and Kilburn DG. (2002). Identification and glucan-binding properties of a new carbohydrate-binding module family. Biochem J. 2002;361(Pt 1):35-40. DOI:10.1042/0264-6021:3610035 |
- Boraston AB, Kwan E, Chiu P, Warren RA, and Kilburn DG. (2003). Recognition and hydrolysis of noncrystalline cellulose. J Biol Chem. 2003;278(8):6120-7. DOI:10.1074/jbc.M209554200 |
- Tsukimoto K, Takada R, Araki Y, Suzuki K, Karita S, Wakagi T, Shoun H, Watanabe T, and Fushinobu S. (2010). Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module. FEBS Lett. 2010;584(6):1205-11. DOI:10.1016/j.febslet.2010.02.027 |
- Notenboom V, Boraston AB, Chiu P, Freelove AC, Kilburn DG, and Rose DR. (2001). Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study. J Mol Biol. 2001;314(4):797-806. DOI:10.1006/jmbi.2001.5153 |
- Boraston AB, Nurizzo D, Notenboom V, Ducros V, Rose DR, Kilburn DG, and Davies GJ. (2002). Differential oligosaccharide recognition by evolutionarily-related beta-1,4 and beta-1,3 glucan-binding modules. J Mol Biol. 2002;319(5):1143-56. DOI:10.1016/S0022-2836(02)00374-1 |
- Araki Y, Karita S, Tanaka A, Kondo M, and Goto M. (2009). Characterization of family 17 and family 28 carbohydrate-binding modules from Clostridium josui Cel5A. Biosci Biotechnol Biochem. 2009;73(5):1028-32. DOI:10.1271/bbb.80802 |
- Fox JM, Jess P, Jambusaria RB, Moo GM, Liphardt J, Clark DS, and Blanch HW. (2013). A single-molecule analysis reveals morphological targets for cellulase synergy. Nat Chem Biol. 2013;9(6):356-61. DOI:10.1038/nchembio.1227 |
- Mosbah A, Tardif C, Bornet O, Valette O, Henrissat B, and Darbon H. (2002). Assignment of the 1H, 13C, and 15N resonances of the 22,5 kDa CBM28 module of the cellulase Cel5I of Clostridium cellulolyticum. J Biomol NMR. 2002;23(2):157-8. DOI:10.1023/a:1016354623398 |
- Jamal S, Nurizzo D, Boraston AB, and Davies GJ. (2004). X-ray crystal structure of a non-crystalline cellulose-specific carbohydrate-binding module: CBM28. J Mol Biol. 2004;339(2):253-8. DOI:10.1016/j.jmb.2004.03.069 |