CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Carbohydrate Binding Module Family 35
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Harry Gilbert^^^
- Responsible Curator: ^^^Wade Abbott^^^
| CAZy DB link | |
| http://www.cazy.org/CBM35.html |
Ligand specificities
CBM35 is predominantly a bacterial family that comprise protein modules of around 140 amino acids that display a variety of different specificities. Some CBM35s that bind to uronic acids (UA-CBM35) recognise only 4,5Δunsaturated deoxyGalA (Pel-CBM35), while other UA-CBM35s can bind to both 4,5Δ-unsaturated deoxyGalA and glucuronic acid (GlcA) [1]. CBM35s that target neutral sugars can bind to the internal regions of β1,4-mannan [2], the non-reducing galactose residues in β1,3-galactan [3], the galactose units linked α1,6 to the backbone of β-mannan [4], and inferred from crystal structural data,α1,6- and α1,4-glucans [5, 6]. All experimentally determined binding was enthalpically driven with affinities varying from 1 to 10 x 103 M-1.
Structural features
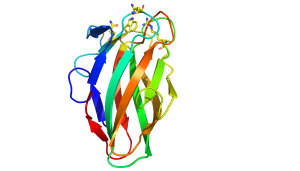
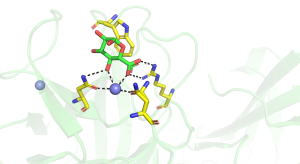
The structures of CBM35s with different specificities have been reported. All these proteins adopt a β-sandwich fold in which the two β-sheets contain four and five antiparallel β-strands, respectively. In one of the glucan binding CBM35s the concave surface of one of the β-sheets; comprises the binding site for a circular tetrasaccharide (see below). In all the other CBM35s, however, the ligand binding site is located in the "Variable Loop Site" [7] that plays a role in connecting the two β-sheets (Figure 1). In the UA-CBM35s a calcium is located in the ligand binding site that interacts via a solvent molecule with the carboxylate (Figure 1 [1]). The Asn ligands for the metal ion also hydrogen bond with the sugar and, by making interactions only with an equatorial O4 select for GlcA over GalA. A conserved arginine makes bidentate polar interactions with the carboxylate and a tryptophan forms parallel hydrophobic interactions with the pyranose ring. Subtle changes in the polar residues in the ligand binding site of a pectate lyase CBM35 confers specificity for only the unsaturated uronic acid. The role of these polar and hydrophobic residues in glycan binding was confirmed by mutagenesis. The pocket topology of the ligand binding site in which only O1 is solvent exposed explains why these CBMs are exo-binding, recognising the non-reducing terminal sugars and are thus classified as type C modules.
The ligand binding site of the CBM35 that binds to β-mannan (Man-CBM35) adopts a cleft topology and, reflecting its endo-binding mode is designated a a type B module [8]. In contrast to the majority of CBMs, which are generally rigid proteins, Man-CBM35 undergoes significant conformational change upon ligand binding. The curvature of the binding site and the narrow binding cleft are likely to be the main determinants of binding specificity. The predicted solvent exposure of O6 at several subsites provides an explanation for the observed accommodation of decorated mannans. Two of the key aromatic residues in Man-CBM35 that interact with mannopentaose are conserved in mannanase-derived CBM35s.
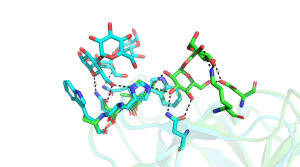
A very large number of structures of CBM35s derived from glucantransferases have been reported [5, 6, 9], with several of these enzymes containing multiple copies of these modules. For example a CBM35 module (BcCBM35-1) is located in the loop connecting the β7 strand and α-helix of the TIM barrel GH66 catalytic module of a Bacillus circulans cycloisomaltooligosaccharide glucanotransferase, while a second CBM35 (BcCBM35-2) in this enzyme is a discrete module [5]. BcCBM35-1 and BcCBM35-2 bind to α1,6- and α1,4-glucans, respectively, in the same region described above for the UA-CBM35s. In these CBMs there is no calcium in the ligand binding site, however, the crucial stacking tryptophan residue is conserved. The ligand binding site of BcCBM35-1 has a pocket-like topology, similar to UA-CBM35s. The pocket, however, lacks the conserved arginine in UA-CBMs that makes polar interactions with the carboxylate of GlcA ligand (Figure 2). The lack of this bulky amino acid creates the necessary space for O6 to be solvent exposed, explaining why the CBM can interact with internal sugars in the highly curved structure of α1,6-glucans. The binding site for α1,4-glucans in CBM35-2 is displaced by 11 Å from the binding pocket in the other CBM35s. In addition to making polar contacts with ligand through residues that are conserved in the other CBM35s, BcCBM35-2 also contains a surface tryptophan and two asparagine residues, absent in other family members, that make strong hydrogen bonds and apolar interactions with the α1,4-glucan (Figure 3). In the CBM35 of the Listeria monocytogenes glucantransferase CAFÉ, an alternating α1,3-α1,6- circular tetrasaccharide binds on the concave surface adopted by one of the β-sheets [9] (PDB ID 5i0d).
Functionalities
Although CBM35 constitutes a discrete protein family, phylogenetic analysis showed that it displays a distant relationship with family 6 CBMs [2]. Based on sequence similarities Family 35 CBMs partitions into four major clades (I-IV). Functional predictions for each subfamily are based upon the distributions of CBM35s with known specificities, including carbohydrates with galacto (I), manno (II), gluco (III), and uronate (IV) configurations [4]. The analysis reveals some enzymes that contain multiple copies of CBM35s that cluster into different subfamilies, illustrated by BcCB35-1 and BcCBM35-2, described above, or are located in the same subfamily suggesting conserved ligand recognition leading to enhanced affinity through avidity effects. A more detailed phylogenetic analysis of CBM35 identified five molecular regions that contribute to specificity and affinity [4].
The functional significance of CBM35s have been discussed in the literature. It has been proposed that the recognition of Δ4,5-GalA by CBM35s that are components of plant cell wall (PCW) degrading enzymes, indicates that these CBMs play a role in targeting their cognate CAZymes to regions of the PCW accessible to biological degradation. Pectins are arguably the most accessible region of the plant cell wall and indeed cloak other polysaccharides, such as xyloglucan, occluding them from enzyme attack [10]. Thus, by targeting enzymes to regions rich in Δ4,5-GalA, the UA-CBM35s are directing their cognate catalytic modules to areas of the PCW that are particularly accessible to further biological degradation. This hypothesis was supported by extensive binding of the UA-CBM35 to the pectin rich primary cell walls of plants only after pectate lyase treatment [1]. It should be emphasized, however, that a group of three UA-CBM35s form cassettes with cellulose binding CBM2s (these CBM2-UA-CBM35 cassettes are identical in amino acid sequence even at the nucleotide level [11, 12]), and are appended to three xylan degrading enzymes (esterase, arabinofuranosidase and endo-xylanase). These UA-CBM35s also display significant affinity for GlcA, a side chain present in several xylans [13]. Thus, this plasticity in uronic acid recognition provides the enzyme with a flexible targeting strategy that is tailored to the plant biomass presented to the bacterium. It was argued that this cohort of CBM35s directed their enzymes to regions of cell walls that are being actively degraded but, as xylan structures are revealed, the enzyme is shuttled onto the hemicellulosic polysaccharide affording the hydrolases access to its target substrate [1]. Indeed appending the UA-CBM35 to a xylanase enhanced its activity against plant cell walls in muro [14].
In addition to their contribution to enzyme action it has been shown that a UA-CBM35, appended to an exo-β-d-glucosaminidase from the soil bacterium Amycolatopsis orientalis, interacts with the cell wall of the organism, which are known to contain GlcA. Immunofluorescence microscopy showed that the UA-CBM functions as an adhesion molecule tethering the exo-β-D-glucosaminidase to the bacterial cell surface [1]. The α-glucan binding CBMs in glucantransferases appear to contribute to substrate binding in concert with the catalytic module. For example BcCBM35-1 appears to contribute the -8 subsite of the enzyme [5]. This hypothesis was supported by mutagenensis of the two sugar binding sites of BcCBM35-1, while led to a substantial increase in the proportion of large cycloisomaltooligosaccharides with a degree of polymerization <9. In addition to influencing enzyme activity, UA-CBM35s have also been used to explore the nature of GlcA in PCW, as these modules bind to GlcA but not 4-Me-O-GlcA [14]
Family Firsts
- First Identified
- The Man-CBM35 and a UA-CBM35 from a C. japonicus GH5 mannanase and GH62 arabinofuranosidase, respectively, were the first members of this family to be identified and characterized [2].
- First Structural Characterization
- The first structural characterization of a CBM35 was Man-CBM35 derived from the C. japonicus endo-mannanase Man5C [8]
References
- Montanier C, van Bueren AL, Dumon C, Flint JE, Correia MA, Prates JA, Firbank SJ, Lewis RJ, Grondin GG, Ghinet MG, Gloster TM, Herve C, Knox JP, Talbot BG, Turkenburg JP, Kerovuo J, Brzezinski R, Fontes CM, Davies GJ, Boraston AB, and Gilbert HJ. (2009). Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc Natl Acad Sci U S A. 2009;106(9):3065-70. DOI:10.1073/pnas.0808972106 |
- Bolam DN, Xie H, Pell G, Hogg D, Galbraith G, Henrissat B, and Gilbert HJ. (2004). X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J Biol Chem. 2004;279(22):22953-63. DOI:10.1074/jbc.M313317200 |
- Ichinose H, Yoshida M, Kotake T, Kuno A, Igarashi K, Tsumuraya Y, Samejima M, Hirabayashi J, Kobayashi H, and Kaneko S. (2005). An exo-beta-1,3-galactanase having a novel beta-1,3-galactan-binding module from Phanerochaete chrysosporium. J Biol Chem. 2005;280(27):25820-9. DOI:10.1074/jbc.M501024200 |
- Correia MA, Abbott DW, Gloster TM, Fernandes VO, Prates JA, Montanier C, Dumon C, Williamson MP, Tunnicliffe RB, Liu Z, Flint JE, Davies GJ, Henrissat B, Coutinho PM, Fontes CM, and Gilbert HJ. (2010). Signature active site architectures illuminate the molecular basis for ligand specificity in family 35 carbohydrate binding module. Biochemistry. 2010;49(29):6193-205. DOI:10.1021/bi1006139 |
- Suzuki N, Fujimoto Z, Kim YM, Momma M, Kishine N, Suzuki R, Suzuki S, Kitamura S, Kobayashi M, Kimura A, and Funane K. (2014). Structural elucidation of the cyclization mechanism of α-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase. J Biol Chem. 2014;289(17):12040-12051. DOI:10.1074/jbc.M114.547992 |
- Fujimoto Z, Suzuki N, Kishine N, Ichinose H, Momma M, Kimura A, and Funane K. (2017). Carbohydrate-binding architecture of the multi-modular α-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces α-1,6-glucosyl-α-glucosaccharides from starch. Biochem J. 2017;474(16):2763-2778. DOI:10.1042/BCJ20170152 |
- Abbott DW and van Bueren AL. (2014). Using structure to inform carbohydrate binding module function. Curr Opin Struct Biol. 2014;28:32-40. DOI:10.1016/j.sbi.2014.07.004 |
- Tunnicliffe RB, Bolam DN, Pell G, Gilbert HJ, and Williamson MP. (2005). Structure of a mannan-specific family 35 carbohydrate-binding module: evidence for significant conformational changes upon ligand binding. J Mol Biol. 2005;347(2):287-96. DOI:10.1016/j.jmb.2005.01.038 |
- Light SH, Cahoon LA, Mahasenan KV, Lee M, Boggess B, Halavaty AS, Mobashery S, Freitag NE, and Anderson WF. (2017). Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity. Structure. 2017;25(2):295-304. DOI:10.1016/j.str.2016.12.007 |
- Marcus SE, Verhertbruggen Y, Hervé C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WG, and Knox JP. (2008). Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8:60. DOI:10.1186/1471-2229-8-60 |
- Kellett LE, Poole DM, Ferreira LM, Durrant AJ, Hazlewood GP, and Gilbert HJ. (1990). Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990;272(2):369-76. DOI:10.1042/bj2720369 |
- Ferreira LM, Wood TM, Williamson G, Faulds C, Hazlewood GP, Black GW, and Gilbert HJ. (1993). A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993;294 ( Pt 2)(Pt 2):349-55. DOI:10.1042/bj2940349 |
- Urbanowicz BR, Peña MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, Foston M, Li H, O'Neill MA, Ragauskas AJ, Darvill AG, Wyman C, Gilbert HJ, and York WS. (2012). 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc Natl Acad Sci U S A. 2012;109(35):14253-8. DOI:10.1073/pnas.1208097109 |