CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Carbohydrate Binding Module Family 44"
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | CBM44 targets ß-1,4-polymers such as xyloglucan and cellulose (hydroxyethylcellulose and Avicel), mixed linkage ß-1,3/ß-1,4- glucans (lichenan and barley) or glucomannan (konjac) <cite>Najmudin2006</cite> and is classified as a type B CBM. Affinity for xylan was very low and binding to laminarin, curdlan, pullulan, pustulan, galactomannan or galactan was negative. Isothermal titration calorimetry (ITC) revealed highest affinity for xyloglucan as a polysaccharide (~81.6 x 10<sup>4</sup> M<sup>-1</sup>), which was comparable to the affinity for cellohexaose as an oligosaccharide (~72.8 x 10<sup>4</sup> M<sup>-1</sup>). For other cello-oligosaccharides, this affinity decreased with decreasing chain length, while no binding was detected for cellotriose <cite>Najmudin2006</cite>. | + | CBM44 from the CtCel9D-Cel44A enzyme produced by ''Acetivibrio thermocellus'' (formerly ''Clostridium thermocellum'' <cite>#Ahsan1996 #Arai2003</cite>) targets ß-1,4-polymers such as xyloglucan and cellulose (hydroxyethylcellulose and Avicel), mixed linkage ß-1,3/ß-1,4- glucans (lichenan and barley) or glucomannan (konjac) <cite>Najmudin2006</cite> and is classified as a type B CBM. Affinity for xylan was very low and binding to laminarin, curdlan, pullulan, pustulan, galactomannan or galactan was negative. Isothermal titration calorimetry (ITC) revealed highest affinity for xyloglucan as a polysaccharide (~81.6 x 10<sup>4</sup> M<sup>-1</sup>), which was comparable to the affinity for cellohexaose as an oligosaccharide (~72.8 x 10<sup>4</sup> M<sup>-1</sup>). For other cello-oligosaccharides, this affinity decreased with decreasing chain length, while no binding was detected for cellotriose <cite>Najmudin2006</cite>. |
== Structural Features == | == Structural Features == | ||
Revision as of 01:40, 10 January 2023
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| CAZy DB link | |
| http://www.cazy.org/CBM44.html |
Ligand specificities
CBM44 from the CtCel9D-Cel44A enzyme produced by Acetivibrio thermocellus (formerly Clostridium thermocellum [1, 2]) targets ß-1,4-polymers such as xyloglucan and cellulose (hydroxyethylcellulose and Avicel), mixed linkage ß-1,3/ß-1,4- glucans (lichenan and barley) or glucomannan (konjac) [3] and is classified as a type B CBM. Affinity for xylan was very low and binding to laminarin, curdlan, pullulan, pustulan, galactomannan or galactan was negative. Isothermal titration calorimetry (ITC) revealed highest affinity for xyloglucan as a polysaccharide (~81.6 x 104 M-1), which was comparable to the affinity for cellohexaose as an oligosaccharide (~72.8 x 104 M-1). For other cello-oligosaccharides, this affinity decreased with decreasing chain length, while no binding was detected for cellotriose [3].
Structural Features
Like many CBMs, CBM44 exhibits a typical ß-sandwich fold: two antiparallel ß-sheets form a concave and a convex surface. The concave surface forms a deep hydrophobic ligand-binding cleft that is estimated to accommodate up to five glucose residues (~24 Å). Here, three tryptophans (W189, W194 and W198) act as key residues to mediate ligand binding (Figure 1), as confirmed by affinity gel electrophoresis (AGE) and ITC analyses of specific mutants [3]. The orientation of the tryptophans corresponds to the slightly twisted conformation of cello-oligosaccharides in solution [4].
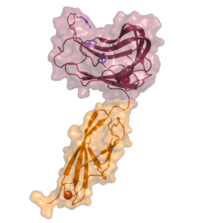
Functionalities
Carbohydrate binding and targeting were described as functions of CBM44. While carbohydrate binding was confirmed by AGE and ITC, the targeting effect was analyzed by the ability of CBM44 to maintain enzyme activity of GH44 (Cel44A) exposed to a substrate mixture [3]. The ensemble of a GH44 and a CBM44 was found in the multimodular cellulase CtCel9D-Cel44A from Acetivibrio thermocellus and ligand spectra of both modules are consistent [3]. In the targeting experiment, the fused GH44-CBM44 construct was compared to the truncated GH44. In separate xyloglucan or carboxymethylcellulose incubations, the activity of GH44-CBM44 was comparable to that of GH44 alone. In a next step, substrate mixtures were used. These mixtures also contained laminarin and pustulan, which were not degraded. While GH44 activity was decreased, activity was restored by GH44-CBM44. Thus, given the complex carbohydrates present in plant cell walls, CBM44 may guide the enzyme to its target sugars. Such effects have also been achieved in synthetic CBM44-enzyme chimeras, e.g. GH12 or GH28 [5, 6].
Family Firsts
- First Identified
- CBM44 was first described as an unknown C-terminal domain in the multimodular cellulase CtCel9D-Cel44A in Acetivibrio thermocellus, formerly designated as CelJ and as Clostridium thermocellum [1, 2]. The protein also features two catalytic domains (GH44 and GH9), a CBM30 and an internal dockerin to target the protein to the cellulosome. The C-terminal region was then identified as having a PKD module as well as a novel CBM binding ß-1,4-glucans, hereby founding the family 44 [3].
- First Structural Characterization
- The same CBM44, together with the preceding PKD module, represents the first structurally characterized candidate of this family (PDB 2C26) [3].
References
- Ahsan MM, Kimura T, Karita S, Sakka K, and Ohmiya K. (1996). Cloning, DNA sequencing, and expression of the gene encoding Clostridium thermocellum cellulase CelJ, the largest catalytic component of the cellulosome. J Bacteriol. 1996;178(19):5732-40. DOI:10.1128/jb.178.19.5732-5740.1996 |
- Arai T, Araki R, Tanaka A, Karita S, Kimura T, Sakka K, and Ohmiya K. (2003). Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J Bacteriol. 2003;185(2):504-12. DOI:10.1128/JB.185.2.504-512.2003 |
- Najmudin S, Guerreiro CI, Carvalho AL, Prates JA, Correia MA, Alves VD, Ferreira LM, Romão MJ, Gilbert HJ, Bolam DN, and Fontes CM. (2006). Xyloglucan is recognized by carbohydrate-binding modules that interact with beta-glucan chains. J Biol Chem. 2006;281(13):8815-28. DOI:10.1074/jbc.M510559200 |
-
Sugiyama H, Hisamichi K, Usui T, Sakai K, Ishiyama J (2000). A study of the conformation of beta-1,4-linked glucose oligomers, cellobiose to cellohexaose, in solution. J Mol Struct. 2000;556(1-3):173-7. DOI: 10.1016/S0022-2860(00)00630-X.
- Furtado GP, Santos CR, Cordeiro RL, Ribeiro LF, de Moraes LA, Damásio AR, Polizeli Mde L, Lourenzoni MR, Murakami MT, and Ward RJ. (2015). Enhanced xyloglucan-specific endo-β-1,4-glucanase efficiency in an engineered CBM44-XegA chimera. Appl Microbiol Biotechnol. 2015;99(12):5095-107. DOI:10.1007/s00253-014-6324-0 |
-
Carli S, Meleiro LP, Salgado JCS, Ward RJ (2022). Synthetic carbohydrate-binding module-endogalacturonase chimeras increase catalytic efficiency and saccharification of lignocellulose residues. Biomass Conv Bioref. 2022. DOI: 10.1007/s13399-022-02716-6.