CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Carbohydrate Binding Module Family 58"
Harry Brumer (talk | contribs) m (Moved Fig. 1 to right) |
Harry Brumer (talk | contribs) m (fixed line spacing at top of page) |
||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | |||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | |||
* [[Author]]: ^^^Nicole Koropatkin^^^ | * [[Author]]: ^^^Nicole Koropatkin^^^ | ||
* [[Responsible Curator]]: ^^^Nicole Koropatkin^^^ | * [[Responsible Curator]]: ^^^Nicole Koropatkin^^^ | ||
---- | ---- | ||
| − | |||
<!-- The data in the table below should be updated by the Author/Curator according to current information on the family --> | <!-- The data in the table below should be updated by the Author/Curator according to current information on the family --> | ||
<div style="float: right"> | <div style="float: right"> | ||
| Line 17: | Line 14: | ||
|} | |} | ||
</div> | </div> | ||
| − | |||
<!-- This is the end of the table --> | <!-- This is the end of the table --> | ||
Revision as of 14:16, 10 September 2017
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Nicole Koropatkin^^^
- Responsible Curator: ^^^Nicole Koropatkin^^^
| CAZy DB link | |
| http://www.cazy.org/CBM58.html |
Ligand specificities
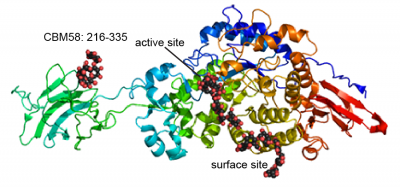
Only a single CBM58 family member has been characterized, the founding member from the neopullulanase SusG of the human gut symbiont Bacteroides thetaiotaomicron (Figure 1) [1]. The crystal structure of SusG featuring CBM58 revealed that this domain binds to maltoheptaose as well as acarbose, a pseudotetrasaccharide amylase inhibitor. Isothermal titration calorimetry with maltoheptaose and α-cyclodextrin as well as isotherm depletion assays with insoluble cornstarch further confirmed that CBM58 is a starch-specific CBM [1]. Based upon these data, the CBM58 family is believed to bind exclusively to α1,4-linked glucan structures in starch.
Structural Features
The crystal structure of CBM58 from SusG of B. thetaiotaomicron displays a canonical β-sandwich fold, with a single binding site accomodated by the loops connecting β3 and β4, β6 and β7, and β7 and β8 [1]. These loops fold over one of the β-sheets. Because CBM58 recognizes internal regions of the polypeptide chain, it can be classified as a type B CBM [2]. Like many starch-specific CBMs [3], the helical α-glucan is cradled by aromatic stacking interactions with two SusG CBM58 aromatic residues, W287 and W299, as well as hydrogen bonding interactions with K304, N330 and Y260 that aid in positioning the adjacent glucose residues that stack with the tryptophans [1].
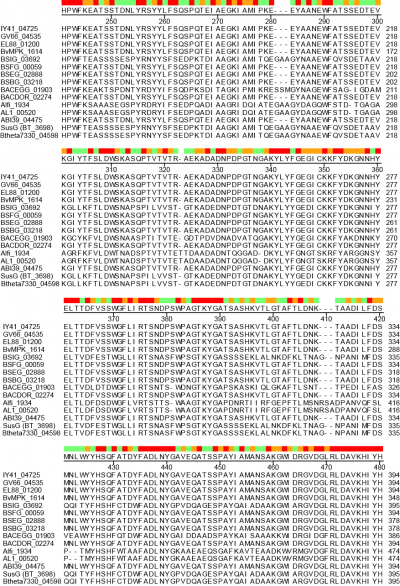
A unique facet of the CBM58 of SusG is its position in the middle of the GH13 amylase fold, occupying residues 216-335 of the protein. This domain is inserted between α3 and β3 of the catalytic B domain (residues 153-215 and 336-363), and essentially expands the typically small B domain of GH13 enzymes. Thus the entire SusG sequence can be described from N- to C-terminus (residue numbers): A domain (43-152) - B domain (153-215) - CBM58 (215-335) - B domain (336-363) - A domain (364-607) - C domain (608-692). In SusG, two short linker sequences from residues 212-217 and 334-338, spanning 12Å between the B domain and CBM58, have B-factors and an amino acid sequence that imply this domain is held in a fixed position [1]. The CBM58 family is small, with the seminal members all arising from the Bacteroidetes phylum. This includes similar GH13 enzymes from isolates of Alistipes finegoldii, Alistipes shahii, Bacteroides dorei, Bacteroides eggerthii and Bacteroides vulgatus. A sequence alignment of 15 representative GH13 enzymes possessing a CBM58 reveals that the location of this domain is invariant as an extension of the B domain (Figure 2). The majority of these CBM58-containing proteins are expressed within canonical polysaccharide utilization loci [4, 5] of the Bacteroidetes that also encode homologs of the TonB-dependent transporter SusC and a homolog of the starch-binding protein SusD.
Functionalities
CBM58 is only found within GH13 enzymes, and the conserved placement of the domain within the B. thetaiotaomicron SusG and like enzymes is unusual as it interrupts the fold of the catalytic domain. In the structure of SusG, the starch-binding face of CBM58 is located 45Å away and on the opposite face of the protein from the catalytic cleft [1]. A mutant enzyme of SusG in which the CBM58 has been deleted retains catalytic activity against small substrates such as PNP-maltohexaose similar to the wild-type enzyme [1]. The CBM-less SusG enzyme is 2-3 fold more active on autoclaved soluble starch such as maize amylopectin, but only retains ~30% of the wild-type activity on insoluble corn starch [1]. B. thetaiotaomicron expressing only an allele for the CBM-less SusG grows the same as wild-type cells on autoclaved soluble starches [6].
The CBM58 of SusG has been replaced with the Halo-tag® protein for single-molecule fluorescence imaging of the SusG protein in live B. thetaiotaomicron cultured on glucose and starch [7]. In these experiments, the SusG-halo fusion partitioned between mobile and confined populations on the cell surface during growth on glucose and starch respectively, and the diffusion coefficient of the protein was measurably slower when interacting with starch granules, and in the presence of the other Sus surface proteins [7].
Family Firsts
- First Identified
- The CBM58 domain was identified during the biochemical and structural characterization of SusG from B. thetaiotaomicron [1].
- First Structural Characterization
- The crystal structure of the GH13 enzyme SusG with maltoheptaose revealed an extended B domain, CBM58, that recognizes starch and maltooligosaccharides [1].
References
- Koropatkin NM and Smith TJ. (2010). SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18(2):200-15. DOI:10.1016/j.str.2009.12.010 |
- Gilbert HJ, Knox JP, and Boraston AB. (2013). Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol. 2013;23(5):669-77. DOI:10.1016/j.sbi.2013.05.005 |
- Machovic M and Janecek S. (2006). Starch-binding domains in the post-genome era. Cell Mol Life Sci. 2006;63(23):2710-24. DOI:10.1007/s00018-006-6246-9 |
- Foley MH, Cockburn DW, and Koropatkin NM. (2016). The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci. 2016;73(14):2603-17. DOI:10.1007/s00018-016-2242-x |
- Grondin JM, Tamura K, Déjean G, Abbott DW, and Brumer H. (2017). Polysaccharide Utilization Loci: Fueling Microbial Communities. J Bacteriol. 2017;199(15). DOI:10.1128/JB.00860-16 |
- Cameron EA, Kwiatkowski KJ, Lee BH, Hamaker BR, Koropatkin NM, and Martens EC. (2014). Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. mBio. 2014;5(5):e01441-14. DOI:10.1128/mBio.01441-14 |
- Karunatilaka KS, Cameron EA, Martens EC, Koropatkin NM, and Biteen JS. (2014). Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts. mBio. 2014;5(6):e02172. DOI:10.1128/mBio.02172-14 |