CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Carbohydrate Binding Module Family 71
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Ben Pluvinage^^^
- Responsible Curator: ^^^Al Boraston^^^
| CAZy DB link | |
| http://www.cazy.org/CBM71.html |
Ligand specificities
The CBM family 71 was created in September 2014 with the characterization of the large multimodular β-galactosidase BgaA from Streptococcus pneumoniae [1]. Two CBMs, CBM71-1 (residues 1463-1645) and CBM71-2 (residues 1828-1998), were identified in the N-terminal region of this protein. The functional characterization of the CBMs reveals a binding specificity limited to lactose (galactopyranosyl-β-1,4-D-glucose) and LacNAc (galactopyranosyl-β-1,4-N-acetyl-D-glucosamine).
Structural Features
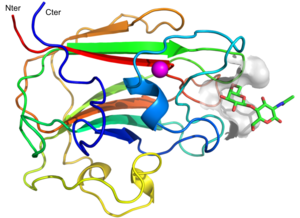
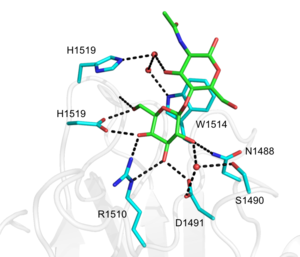
The structure of CBM71-1 [1] solved by X-ray crystallography shows a β-sandwich fold comprising opposing sheets of 4- and 5-antiparallel β-strands and a bound structural metal ion modelled as a calcium (Figure 1). CBM71-1 structure in complex with LacNAc reveals a shallow binding site located at the apex of the β-fold opposite the N- and C-termini, making CBM71s type C CBMs [2]. The basis of CBM71-1 specificity for sugars with a terminal galactose resides in the W1514 side chain configuration, which provides CH-pi interactions with both β-linked pyranose rings of the disaccharide (Figure 2). CBM71-2, which possesses 35% sequence identity with CBM71-1, presents a very similar fold and an almost identical binding site [1].
Functionalities
The CBM71s are found as ancillary modules in the β-galactosidase BgaA from S. pneumoniae [1]. BgaA is a large multimodular cell surface exposed CAZyme comprising 17 modules of 7 different types. However, only the catalytic module belonging to the family 2 glycoside hydrolase [3] has been fully characterized along with the characterization of CBM71s [1]. In addition to their classical CBM role of focusing the enzyme to its substrate, the streptococcal CBM71s have been shown to contribute to pneumococcal adherence by binding lactose- and LacNAc-containing cell surface glycoconjugates [1].
Family Firsts
- First Identified
- The CBM71 modules were first identified through the characterization of the β-galactosidase BgaA from S. pneumoniae [1]. CBM71-1 and CBM71-2 are the founding, and only characterized, members of the family.
- First Structural Characterization
- The first crystal structures of family CBM71s were from the streptococcal β-galactosidase BgaA [1]. The crystallographic structures of a seleno-methionine derivative of CBM71-1 (PDB ID 4CUA), CBM71-1 in complex with LacNAc (PDB ID 4CUB) and CBM71-2 (PDB ID 4CU9) were deposited in the Protein Data Bank in September 2014.
References
- Singh AK, Pluvinage B, Higgins MA, Dalia AB, Woodiga SA, Flynn M, Lloyd AR, Weiser JN, Stubbs KA, Boraston AB, and King SJ. (2014). Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae β-galactosidase, BgaA. PLoS Pathog. 2014;10(9):e1004364. DOI:10.1371/journal.ppat.1004364 |
- Boraston AB, Bolam DN, Gilbert HJ, and Davies GJ. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769-81. DOI:10.1042/BJ20040892 |
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, and Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233-8. DOI:10.1093/nar/gkn663 |