CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Carbohydrate Binding Module Family 78
Ligand specificities
CBM78 is a family identified in the Ruminococcus flavefaciens cellulosome, a ruminal cellulolytic bacterium (1). CBM78 is a component of an enzyme that contains a catalytic module derived from GH5_4 and displays specificity for β-1,4- and mixed linked β-1,3-1,4-glucans. CBM78 also binds galactomannan and contains a GH26 “β1,4-mannanase” catalytic module (2). CBM78GH5 displays highest affinity for xyloglucan. The similar affinity of CBM78GH5 for cellohexaose and cellopentaose suggests five dominant sugar binding sites. The higher affinity of CBM78GH5 for XXXG than Cellotetraose, suggests a recognition of the xylose side chains. No binding to regenerated (noncrystalline) insoluble cellulose (RC) is detected.
Structural Features
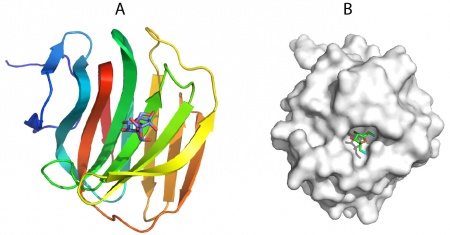
The structure of CBM78RfGH5 was solved using single-wavelength anomalous diffraction (SAD) methods and selenomethionyl protein to a resolution of 2.0 Å. CBM78RfGH5 has a β-sandwich fold and contains two β-sheets, 1 and 2, respectively (Figure 1) (2). β-sheet 2 forms a cleft in which aromatic residues are a dominant feature. Trp496, Trp554, Tyr555, and Phe479 are aligned along the cleft. This hydrophobic region is the glucan binding site in CBM78RfGH5 (2).
Functionalities
CBM78 plays an enzyme-targeting role that is specific to Ruminococcus and a contribution to enzyme function in a highly complex scaffolding. CBM78 that binds β-glucans is component of enzyme that contains catalytic modules derived from GH5_4 with endo-β1,4-glucanases activity. CBM78 is also component of enzyme that contains catalytic modules derived from GH26 with β1,4-mannanase activity. Mutagenesis experiments confirmed the importance of the aromatic residues in ligand recognition of CBM78RfGH5. Alanine substitution of Trp496 or Trp554 in CBM78RfGH5, which are conserved in the CBM family, resulted in complete loss of binding to all ligands. The mutants F479A and Y555A bound to xyloglucan, but not to barley β-glucan or HEC.The variant Q552A recognized xyloglucan and barley β-glucan, but not HEC. No binding to regenerated (noncrystalline) insoluble cellulose is detected and it is explained by the narrow binding cleft of CBM78RfGH5.The mutagenesis data show that different residues play distinct roles in ligand recognition, explaining why this CBM can bind to a range of β-glucans.
Family Firsts
- First Identified
CBM78 from the Ruminococcus flavefaciens CBM78RfGH5 and CBM78RfGH26 .
- First Structural Characterization
The first available crystal structure and the first complex structure of a CBM78 is from CBM78RfGH5.
References
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, and Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233-8. DOI:10.1093/nar/gkn663 |
-
Davies, G.J. and Sinnott, M.L. (2008) Sorting the diverse: the sequence-based classifications of carbohydrate-active enzymes. The Biochemist, vol. 30, no. 4., pp. 26-32. Download PDF version.
- Boraston AB, Bolam DN, Gilbert HJ, and Davies GJ. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769-81. DOI:10.1042/BJ20040892 |
- Hashimoto H (2006). Recent structural studies of carbohydrate-binding modules. Cell Mol Life Sci. 2006;63(24):2954-67. DOI:10.1007/s00018-006-6195-3 |
- Shoseyov O, Shani Z, and Levy I. (2006). Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev. 2006;70(2):283-95. DOI:10.1128/MMBR.00028-05 |
- Guillén D, Sánchez S, and Rodríguez-Sanoja R. (2010). Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol. 2010;85(5):1241-9. DOI:10.1007/s00253-009-2331-y |
- Armenta S, Moreno-Mendieta S, Sánchez-Cuapio Z, Sánchez S, and Rodríguez-Sanoja R. (2017). Advances in molecular engineering of carbohydrate-binding modules. Proteins. 2017;85(9):1602-1617. DOI:10.1002/prot.25327 |