CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Carbohydrate Binding Module Family 78
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| http://www.cazy.org/CBM78.html |
Ligand specificities
CBM78 is a family identified in the ruminal cellulolytic bacterium Ruminococcus flavefaciens [1] cellulosome (a lignocellulose degrading multienzyme complex; see [2] for a review of cellulosomes). The only characterized CBM78 modules are CBM78RfGH5 and CBM78RfGH26, which are components of enzymes containing catalytic modules derived from GH5_4 and GH26, respectively. While the ligand binding of CBM78RfGH5 was quantified by isothermal titration calorimetry, the specificity of CBM78RfGH26 was determined semi-quantitatively using microarrays [3]. Both CBM78 modules bind to β-1,4- and mixed linked β-1,3-1,4-glucans, however, CBM78RfGH26, also binds galactomannan and contains a GH26 "β1,4-mannanase" catalytic module [3]. CBM78RfGH5 displays a higher affinity for xyloglucan relative to cellulose ligands. The similar affinity of CBM78RfGH5 for cellohexaose and cellopentaose suggests five dominant sugar binding sites. The higher affinity of CBM78RfGH5 for the xyloglucan derived oligosaccharide XXXG (where X is a backbone glucose decorated with xylose and G is an unsubstituted glucose) relative to cellotetraose suggests recognition of the xylose side chains. No binding to regenerated (noncrystalline) insoluble cellulose (RC) was detected. Isothermal titration calorimetry showed that ligand binding was enthalpically driven and coverage of polysaccharides at saturation indicated an endo-mode of binding. CBM78 is thus a type B CBM family.
Structural Features
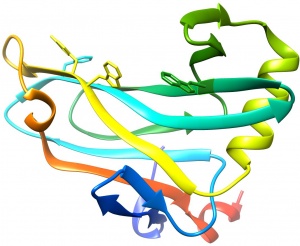
The structure of CBM78RfGH5 was solved using single-wavelength anomalous diffraction (SAD) methods and selenomethionyl protein to a resolution of 2.0 Å. CBM78RfGH5 has a β-sandwich fold and contains two β-sheets, 1 and 2, respectively (Figure 1) [3]. β-sheet 2 forms a cleft in which aromatic residues are a dominant feature, typical of many type B CBM families (see [4] for review). Trp496, Trp554, Tyr555, and Phe479 are aligned along the cleft. This hydrophobic region is the glucan binding site in CBM78RfGH5 [3].
Functionalities
CBM78 modules play an enzyme-targeting role that is specific to the Ruminococcus, which has a particularly complex repertoire of scaffoldins (cellulosome assembly proteins) [5]. The specificity of CBM78RfGH5 for β-glucans is consistent with the endo-β1,4-glucanase activity of the cognate GH5_4 catalytic module [6]. Similarly, the capacity of CBM78RfGH5 to bind β-mannan is consistent with the appended GH26 catalytic module, a family in which the characterized enzymes are predominantly β1,4-mannanases (e.g. [7, 8] and see [9, 10] for reviews). Mutagenesis experiments [3] confirmed the importance of the aromatic residues in ligand recognition of CBM78RfGH5. Alanine substitution of Trp496 or Trp554 in CBM78RfGH5, which are conserved in the CBM family, resulted in complete loss of binding to all ligands. The mutants F479A and Y555A bound to xyloglucan, but not to barley β-glucan or hydroxyethylcellulose (HEC). The variant Q552A recognized xyloglucan and barley β-glucan, but not HEC. No binding to regenerated (noncrystalline) insoluble cellulose was detected consistent with the narrow binding cleft of CBM78RfGH5 [3].The mutagenesis data show that different residues play distinct roles in ligand recognition, explaining why this CBM can bind to a range of β-glucans.
Family Firsts
- First Identified
- CBM78 from the Ruminococcus flavefaciens CBM78RfGH5 and CBM78RfGH26 [3].
- First Structural Characterization
- The first available crystal structure and the first complex structure of a CBM78 is from CBM78RfGH5 [3].
References
- Rincon MT, Dassa B, Flint HJ, Travis AJ, Jindou S, Borovok I, Lamed R, Bayer EA, Henrissat B, Coutinho PM, Antonopoulos DA, Berg Miller ME, and White BA. (2010). Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium, Ruminococcus flavefaciens FD-1. PLoS One. 2010;5(8):e12476. DOI:10.1371/journal.pone.0012476 |
- Fontes CM and Gilbert HJ. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem. 2010;79:655-81. DOI:10.1146/annurev-biochem-091208-085603 |
- Venditto I, Luis AS, Rydahl M, Schückel J, Fernandes VO, Vidal-Melgosa S, Bule P, Goyal A, Pires VM, Dourado CG, Ferreira LM, Coutinho PM, Henrissat B, Knox JP, Baslé A, Najmudin S, Gilbert HJ, Willats WG, and Fontes CM. (2016). Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition. Proc Natl Acad Sci U S A. 2016;113(26):7136-41. DOI:10.1073/pnas.1601558113 |
- Boraston AB, Bolam DN, Gilbert HJ, and Davies GJ. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769-81. DOI:10.1042/BJ20040892 |
- Bensoussan L, Moraïs S, Dassa B, Friedman N, Henrissat B, Lombard V, Bayer EA, and Mizrahi I. (2017). Broad phylogeny and functionality of cellulosomal components in the bovine rumen microbiome. Environ Microbiol. 2017;19(1):185-197. DOI:10.1111/1462-2920.13561 |
- Aspeborg H, Coutinho PM, Wang Y, Brumer H 3rd, and Henrissat B. (2012). Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol. 2012;12:186. DOI:10.1186/1471-2148-12-186 |
- Hogg D, Pell G, Dupree P, Goubet F, Martín-Orúe SM, Armand S, and Gilbert HJ. (2003). The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem J. 2003;371(Pt 3):1027-43. DOI:10.1042/BJ20021860 |
- Bågenholm V, Reddy SK, Bouraoui H, Morrill J, Kulcinskaja E, Bahr CM, Aurelius O, Rogers T, Xiao Y, Logan DT, Martens EC, Koropatkin NM, and Stålbrand H. (2017). Galactomannan Catabolism Conferred by a Polysaccharide Utilization Locus of Bacteroides ovatus: ENZYME SYNERGY AND CRYSTAL STRUCTURE OF A β-MANNANASE. J Biol Chem. 2017;292(1):229-243. DOI:10.1074/jbc.M116.746438 |
- Gilbert HJ, Stålbrand H, and Brumer H. (2008). How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol. 2008;11(3):338-48. DOI:10.1016/j.pbi.2008.03.004 |
- Gilbert HJ (2010). The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010;153(2):444-55. DOI:10.1104/pp.110.156646 |