CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Carbohydrate Binding Module Family 79"
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
CBM79 is a family identified in the ''Ruminococcus flavefaciens'' cellulosome, an anaerobic, cellulolytic bacterium that plays an important role in the ruminal digestion of plant cell walls <cite>RinconMT2010</cite>. Two CBM79s (CBM79-1<sub>RfGH9</sub> and CBM79-2<sub>RfGH9</sub>) were identified in an enzyme that contains a catalytic module derived from [[GH9]] with endo-β-1,4-glucanase activity <cite>VendittoI2016</cite>. Both CBM79s bind to a range of β-1,4- and mixed linked β-1,3-1,4-glucans <cite>VendittoI2016</cite>. The ligand binding of CBM79-1<sub>RfGH9</sub> was quantified by Isothermal Titration Calorimetry (ITC) and semi-quantified using Microarrays <cite>VendittoI2016</cite>. CBM79-1<sub>RfGH9</sub> binds barley β-glucan and hydroxyethylcellulose (HEC) with similar affinities of 10<sup>4</sup> M<sup>-1</sup> <cite>VendittoI2016</cite>. The small increase in ''K''<sub>A</sub> from cellotetraose to cellohexaose | CBM79 is a family identified in the ''Ruminococcus flavefaciens'' cellulosome, an anaerobic, cellulolytic bacterium that plays an important role in the ruminal digestion of plant cell walls <cite>RinconMT2010</cite>. Two CBM79s (CBM79-1<sub>RfGH9</sub> and CBM79-2<sub>RfGH9</sub>) were identified in an enzyme that contains a catalytic module derived from [[GH9]] with endo-β-1,4-glucanase activity <cite>VendittoI2016</cite>. Both CBM79s bind to a range of β-1,4- and mixed linked β-1,3-1,4-glucans <cite>VendittoI2016</cite>. The ligand binding of CBM79-1<sub>RfGH9</sub> was quantified by Isothermal Titration Calorimetry (ITC) and semi-quantified using Microarrays <cite>VendittoI2016</cite>. CBM79-1<sub>RfGH9</sub> binds barley β-glucan and hydroxyethylcellulose (HEC) with similar affinities of 10<sup>4</sup> M<sup>-1</sup> <cite>VendittoI2016</cite>. The small increase in ''K''<sub>A</sub> from cellotetraose to cellohexaose | ||
| − | (KA 4.2 x 10<sup>3</sup> M<sup>-1</sup> for cellotetraose, KA 7.0 x 10<sup>3</sup> M<sup>-1</sup> for cellopentaose and KA 4.9 x 10<sup>3</sup> M<sup>-1</sup> for cellohexaose) suggests that ligand recognition is dominated by four sugar binding sites <cite>VendittoI2016</cite>. The binding to xyloglucan is weaker than the other β-glucans, indicating that the protein cannot recognize the xylose side chains <cite>VendittoI2016</cite>. CBM79-1<sub>RfGH9</sub> binds regenerated (noncrystalline) insoluble cellulose (RC) with a K<sub>A</sub> of 4.8 x 10<sup>4</sup> M<sup>-1</sup> <cite>VendittoI2016</cite>. The ligand binding site located at the concave surface of the protein forms an unusually solvent exposed cleft or planar surface that anyway indicates that CBM79-1<sub>RfGH9</sub> is a type B CBM. | + | (KA 4.2 x 10<sup>3</sup> M<sup>-1</sup> for cellotetraose, KA 7.0 x 10<sup>3</sup> M<sup>-1</sup> for cellopentaose and KA 4.9 x 10<sup>3</sup> M<sup>-1</sup> for cellohexaose) suggests that ligand recognition is dominated by four sugar binding sites <cite>VendittoI2016</cite>. The binding to xyloglucan is weaker than the other β-glucans, indicating that the protein cannot recognize the xylose side chains <cite>VendittoI2016</cite>. CBM79-1<sub>RfGH9</sub> binds regenerated (noncrystalline) insoluble cellulose (RC) with a K<sub>A</sub> of 4.8 x 10<sup>4</sup> M<sup>-1</sup> <cite>VendittoI2016</cite>. The ligand binding site located at the concave surface of the protein forms an unusually solvent exposed cleft or planar surface that anyway indicates that CBM79-1<sub>RfGH9</sub> is a type B CBM <cite>VendittoI2016</cite>. |
== Structural Features == | == Structural Features == | ||
[[File:CBM79-1.jpg|thumb|300px|right|'''Figure 1.''' Crystal structure of CBM79-1<sub>RfGH9</sub>. ([{{PDBlink}}4V1L PDB ID 4V1L])([{{PDBlink}}4V1K PDB ID 4V1K]). The aromatic residues that contribute to ligand recognition are shown.]] | [[File:CBM79-1.jpg|thumb|300px|right|'''Figure 1.''' Crystal structure of CBM79-1<sub>RfGH9</sub>. ([{{PDBlink}}4V1L PDB ID 4V1L])([{{PDBlink}}4V1K PDB ID 4V1K]). The aromatic residues that contribute to ligand recognition are shown.]] | ||
| − | The structure of CBM79-1<sub>RfGH9</sub> was solved using single-wavelength anomalous diffraction (SAD) methods and selenomethionyl protein to a resolution of 1.8 Å <cite>VendittoI2016</cite>. CBM79-1<sub>RfGH9</sub> displays a beta-sandwich fold in which the 12 antiparallel β-strands are organized in two β-sheets 1 and 2 (Figure 1) <cite>VendittoI2016</cite>. β-sheet 2 forms a cleft in which aromatic residues are a dominant feature <cite>VendittoI2016</cite>. Tyr563, Trp564, Tyr597, Trp606 and Trp607 in CBM79-1<sub>RfGH9</sub> form a twisted hydrophobic platform | + | The structure of CBM79-1<sub>RfGH9</sub> was solved using single-wavelength anomalous diffraction (SAD) methods and selenomethionyl protein to a resolution of 1.8 Å <cite>VendittoI2016</cite>. CBM79-1<sub>RfGH9</sub> displays a beta-sandwich fold in which the 12 antiparallel β-strands are organized in two β-sheets 1 and 2 (Figure 1) <cite>VendittoI2016</cite>. β-sheet 2 forms a cleft in which aromatic residues are a dominant feature <cite>VendittoI2016</cite>. Tyr563, Trp564, Tyr597, Trp606 and Trp607 in CBM79-1<sub>RfGH9</sub> form a twisted hydrophobic platform <cite>VendittoI2016</cite>. Two tryptophan residues (Trp564 and Trp606) play a key role in ligand recognition adopting a planar orientation in CBM79-1<sub>RfGH9</sub> <cite>VendittoI2016</cite>. |
== Functionalities == | == Functionalities == | ||
Revision as of 06:43, 28 August 2018
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Immacolata Venditto^^^
- Responsible Curator: ^^^Harry Gilbert^^^
| CAZy DB link | |
| http://www.cazy.org/CBM79.html |
Ligand specificities
CBM79 is a family identified in the Ruminococcus flavefaciens cellulosome, an anaerobic, cellulolytic bacterium that plays an important role in the ruminal digestion of plant cell walls [1]. Two CBM79s (CBM79-1RfGH9 and CBM79-2RfGH9) were identified in an enzyme that contains a catalytic module derived from GH9 with endo-β-1,4-glucanase activity [2]. Both CBM79s bind to a range of β-1,4- and mixed linked β-1,3-1,4-glucans [2]. The ligand binding of CBM79-1RfGH9 was quantified by Isothermal Titration Calorimetry (ITC) and semi-quantified using Microarrays [2]. CBM79-1RfGH9 binds barley β-glucan and hydroxyethylcellulose (HEC) with similar affinities of 104 M-1 [2]. The small increase in KA from cellotetraose to cellohexaose (KA 4.2 x 103 M-1 for cellotetraose, KA 7.0 x 103 M-1 for cellopentaose and KA 4.9 x 103 M-1 for cellohexaose) suggests that ligand recognition is dominated by four sugar binding sites [2]. The binding to xyloglucan is weaker than the other β-glucans, indicating that the protein cannot recognize the xylose side chains [2]. CBM79-1RfGH9 binds regenerated (noncrystalline) insoluble cellulose (RC) with a KA of 4.8 x 104 M-1 [2]. The ligand binding site located at the concave surface of the protein forms an unusually solvent exposed cleft or planar surface that anyway indicates that CBM79-1RfGH9 is a type B CBM [2].
Structural Features
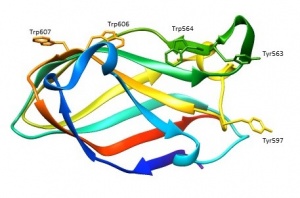
The structure of CBM79-1RfGH9 was solved using single-wavelength anomalous diffraction (SAD) methods and selenomethionyl protein to a resolution of 1.8 Å [2]. CBM79-1RfGH9 displays a beta-sandwich fold in which the 12 antiparallel β-strands are organized in two β-sheets 1 and 2 (Figure 1) [2]. β-sheet 2 forms a cleft in which aromatic residues are a dominant feature [2]. Tyr563, Trp564, Tyr597, Trp606 and Trp607 in CBM79-1RfGH9 form a twisted hydrophobic platform [2]. Two tryptophan residues (Trp564 and Trp606) play a key role in ligand recognition adopting a planar orientation in CBM79-1RfGH9 [2].
Functionalities
CBM79 fulfills an enzyme-targeting role that is specific to Ruminococcus [3]. The specificity of CBM79-1RfGH9 for β-glucans is consistent with endo-β1,4-glucanase activity of the GH9 catalytic module [2]. Mutagenesis experiments confirmed the importance of the aromatic residues in ligand recognition [2]. Alanine substitution of Trp606 in CBM79-1RfGH9, which is conserved in the CBM family, resulted in complete loss of binding to all ligands [2]. The mutants W564A and W607A retained affinity for barley β-glucan but did not bind xyloglucan [2]. The planar topology of the binding cleft of CBM79-1RfGH9 indicated that this protein may interact with components of insoluble cellulose [2]. ITC and pull down assays showed that CBM79-1RfGH9 binds RC [2]. Alanine substitution of W564 and W606 resulted in loss of binding to RC [2]. The importance of conformation of conserved aromatic residues on CBM specificity is evident in family 2 CBMs that bind to cellulose or xylan [4].
Family Firsts
- First Identified
- CBM79 from the Ruminococcus flavefaciens CBM79_1RfGH9 and CBM79_2RfGH9 [2].
- First Structural Characterization
- The first available crystal structure and the first complex structure of a CBM79 is from CBM79_1RfGH9 [2].
References
- Rincon MT, Dassa B, Flint HJ, Travis AJ, Jindou S, Borovok I, Lamed R, Bayer EA, Henrissat B, Coutinho PM, Antonopoulos DA, Berg Miller ME, and White BA. (2010). Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium, Ruminococcus flavefaciens FD-1. PLoS One. 2010;5(8):e12476. DOI:10.1371/journal.pone.0012476 |
- Venditto I, Luis AS, Rydahl M, Schückel J, Fernandes VO, Vidal-Melgosa S, Bule P, Goyal A, Pires VM, Dourado CG, Ferreira LM, Coutinho PM, Henrissat B, Knox JP, Baslé A, Najmudin S, Gilbert HJ, Willats WG, and Fontes CM. (2016). Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition. Proc Natl Acad Sci U S A. 2016;113(26):7136-41. DOI:10.1073/pnas.1601558113 |
- Bensoussan L, Moraïs S, Dassa B, Friedman N, Henrissat B, Lombard V, Bayer EA, and Mizrahi I. (2017). Broad phylogeny and functionality of cellulosomal components in the bovine rumen microbiome. Environ Microbiol. 2017;19(1):185-197. DOI:10.1111/1462-2920.13561 |
- Simpson PJ, Xie H, Bolam DN, Gilbert HJ, and Williamson MP. (2000). The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J Biol Chem. 2000;275(52):41137-42. DOI:10.1074/jbc.M006948200 |