CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Carbohydrate Binding Module Family 81
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Marcelo Liberato^^^
- Responsible Curator: ^^^Fabio Squina^^^
| CAZy DB link | |
| http://www.cazy.org/CBM81.html |
Ligand specificities
The family CBM81 was first described on September 12, 2016 [1]. According to CAZy, another two members have been characterized in the literature since then. The founding CBM81 (named CBM_E1) was identified from a sugar cane soil metagenome library [2], as part of a GH5 endoglucanase, where the catalytic module and the CBM are connected by a 32 amino acids (serine-rich) linker. The CBM_E1 interaction with soluble ligands was determined by Isothermal Titration Calorimetry, resulting in the highest affinity with barley β-glucan (KA of 1.4 x 10-4 M-1), followed by cellohexaose (1.2 x 10-4 M-1), xyloglucan (0.5 x 10-4 M-1) and cellopentaose (0.4 x 10-4 M-1). The protein did not show any affinity for xylan and oligosaccharides such as cellotetraose, mannohexaose, and xylohexaose. The thermodynamic parameters indicated that the CBM_E1 binding to ligands is enthalpically driven, which is a typical characteristic of type B CBMs [3]. On the other hand, based on pull-down assays with insoluble carbohydrates, the CBM_E1 was able to bind to Avicel, but not to Bacterial Microcrystalline Cellulose (BMCC), which is characteristic of type A CBMs. The Avicel is composed of about 40% of amorphous regions [4], these disordered regions of the polysaccharide are the probable CBM_E1 binding region.
Structural Features
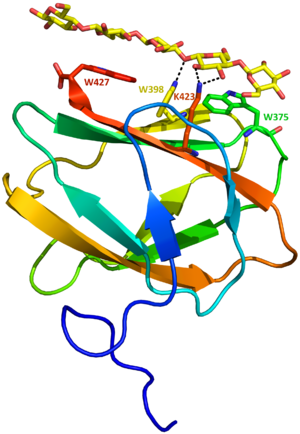
Two structures of CBM81 are deposited in the Protein Data Bank so far. Both from CBM_E1, in the absence (PDB ID 5KLC) and presence of cellopentaose (PDB ID 5KLE) [1]. The structures were solved independent of the catalytic module. CBM_E1 is composed of a beta sandwich with four and five beta-strands connected by loops (Figure 1). Parallel to the beta-strands, three tryptophan are exposed to solvent forming a planar surface, which coordinates ligand binding. No differences were observed between apo and ligand complexed structures.
The CBM81 is the first CBM family to exhibit mixed characteristics from type A and type B CBM binding sites. Type A CBMs bind to the surface of crystalline polysaccharides (such as cellulose and chitin) through CH-pi interactions between the surface aromatic residues and the monosaccharide units [5]. Although the planar surface of this CBM81 member is composed of aromatic residues, similar to type A CBMs, in the crystal structure of the complex one of the tryptophans has the indole ring perpendicular to the oligosaccharide chain, leading to a hydrogen bond instead of a hydrophobic interaction. This observation explains the enthalpically driven binding between the CBM and the ligand and defines the classification of CBM_E1 as a type B.
Functionalities
The CBM_E1 was demonstrated to bind amorphous regions of cellulose, as well as beta-glucan, xyloglucan and cello-oligosaccharides. All these ligands are typical substrates of endoglucanases, which are the enzymes linked to the CBM81 members so far deposited in CAZy. The CBM81 may enhance endoglucanase activity by approximating the catalytic domain to the substrate [6]; however, this effect has not been experimentally demonstrated for this family.
Family Firsts
- First Identified
- The CBM81 CBM_E1 was identified as part of a GH5 endoglucanase, originating from an uncultured microorganism (metagenomics) [1].
- First Structural Characterization
- The first crystal structures of the family CBM81 were from CBM_E1, in absence and presence of the ligand cellopentaose [1].
References
- Campos BM, Liberato MV, Alvarez TM, Zanphorlin LM, Ematsu GC, Barud H, Polikarpov I, Ruller R, Gilbert HJ, Zeri AC, and Squina FM. (2016). A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties. J Biol Chem. 2016;291(45):23734-23743. DOI:10.1074/jbc.M116.744383 |
- Alvarez TM, Paiva JH, Ruiz DM, Cairo JP, Pereira IO, Paixão DA, de Almeida RF, Tonoli CC, Ruller R, Santos CR, Squina FM, and Murakami MT. (2013). Structure and function of a novel cellulase 5 from sugarcane soil metagenome. PLoS One. 2013;8(12):e83635. DOI:10.1371/journal.pone.0083635 |
- Georgelis N, Yennawar NH, and Cosgrove DJ. (2012). Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc Natl Acad Sci U S A. 2012;109(37):14830-5. DOI:10.1073/pnas.1213200109 |
- Hall M, Bansal P, Lee JH, Realff MJ, and Bommarius AS. (2010). Cellulose crystallinity--a key predictor of the enzymatic hydrolysis rate. FEBS J. 2010;277(6):1571-82. DOI:10.1111/j.1742-4658.2010.07585.x |
- Gilbert HJ, Knox JP, and Boraston AB. (2013). Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol. 2013;23(5):669-77. DOI:10.1016/j.sbi.2013.05.005 |
- Hervé C, Rogowski A, Blake AW, Marcus SE, Gilbert HJ, and Knox JP. (2010). Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A. 2010;107(34):15293-8. DOI:10.1073/pnas.1005732107 |