CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Carbohydrate Binding Module Family 87
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| http://www.cazy.org/CBM87.html |
Ligand specificities
The CBM87 family was formed in 2020 after the structural and functional characterization of Agd3, a galactosaminogalactan deacetylase involved in Aspergillus fumigatus biofilm formation [1]. This study also resulted in the creation of a novel carbohydrate esterase superfamily, the CE18 family. Functional characterization has focused on the CBM87 module Agd3CBM87. Binding of Agd3CBM87 to soluble galactosaminogalactan from culture supernatants of A. fumigatus was demonstrated using an ELISA based assay [1]. Galactosaminogalactan is a linear heteropolymer of ⍺-1,4-linked D-galactose (Gal) and partially deacetylated N-acetyl-D-galactosamine (GalNAc and GalN) [2, 3]. Using electrospray ionization mass spectrometry (ESI-MS) and synthesized oligosaccharides Agd3CBM87 was found to have higher affinity for ⍺-1,4-(GalN-⍺-1,4-GalNAc)3 as compared to Gal, GalNAc, or GalN homopolymers of the same linkage [1].
Structural Features
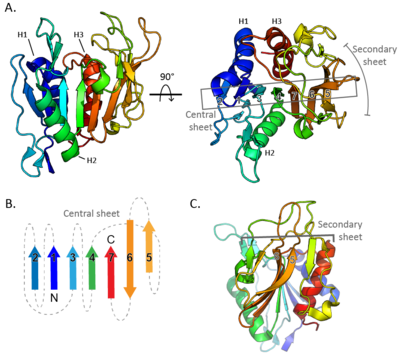
CBM87 members are roughly 220 residues in length and are found N-terminal to CE18 domains. The structure of A. fumigatus Agd3CBM87, was determined using X-ray crystallography to 2.6 Å resolution (PDB 6NWZ) [1]. The structure contains a Rossman-like fold (Figure 1A). The central β-sheet consists of seven strands, with the sixth stand running anti-parallel to the other stands (Figure 1B). The fifth and sixth strands rotate at one end to lie almost perpendicular to the primary central sheet and form the centre of a smaller secondary sheet (Figure 1C). The loops at the C-terminal end of the central β-sheet form a cleft that is predicted to be the location of GAG binding (Figure 2A). Surface exposed conserved aromatic residues (W292 and W344 in A. fumigatus Agd3) were found to contribute to GAG binding (Figure 2B). Binding studies of site directed alanine mutants of W292 and W344 had reduced affinity for GAG produced by A. fumigatus cultures, supporting this hypothesis [1]. The presence of an open binding cleft suggests that CBM87s can bind within a polysaccharide chain and thus may be type B CBMs.
Functionalities
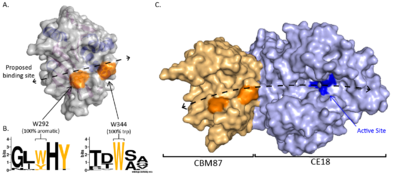
CBM87 members are found N-terminal to CE18 domains (CE18 page of the CAZy Database). The first characterized CE18, Agd3, was found to be important for exopolysaccharide processing and biofilm formation in the fungus A. fumigatus [3]. The structure of Agd3 revealed that the CBM87 domain extends the CE18 active site cleft (Figure 2C). When the CBM87 domain was removed from Agd3, the constructs could not be successfully expressed or purified suggesting that the CBM was required for stability of the catalytic domain [1]. Due to this, the levels of deacetylation activity could not be compared between the CE18 alone and in the presence of the CBM. Therefore, mutations were made in the CBM to disrupt predicted carbohydrate binding sites. Mutation of the conserved surface exposed aromatics, W292 or W344, to alanine led to decreased activity in a GAG deacetylation assay [1]. The CBM87 domain is proposed to contribute important substrate binding sites leading to increase substrate affinity. This was confirmed using mass spectrometry of synthesized hepta-saccharide substrates which revealed a decreased rate of primary deacetylation by the W344A variant. Wild-type Agd3 is capable of deacetylating GAG polymers on multiple subsites [1]. Whether other CBM87 members have similar roles in their cognate systems is still to be determined.
Family Firsts
- First Identified
- The first CBM87 member (Agd3CBM87) to be identified was from A. fumigatus Agd3 [1].
- First Structural Characterization
- The first crystal structure of a CBM87 was Agd3CBM87 [1].
References
- Bamford NC, Le Mauff F, Van Loon JC, Ostapska H, Snarr BD, Zhang Y, Kitova EN, Klassen JS, Codée JDC, Sheppard DC, and Howell PL. (2020). Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation. Nat Commun. 2020;11(1):2450. DOI:10.1038/s41467-020-16144-5 |
- Fontaine T, Delangle A, Simenel C, Coddeville B, van Vliet SJ, van Kooyk Y, Bozza S, Moretti S, Schwarz F, Trichot C, Aebi M, Delepierre M, Elbim C, Romani L, and Latgé JP. (2011). Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 2011;7(11):e1002372. DOI:10.1371/journal.ppat.1002372 |
- Lee MJ, Geller AM, Bamford NC, Liu H, Gravelat FN, Snarr BD, Le Mauff F, Chabot J, Ralph B, Ostapska H, Lehoux M, Cerone RP, Baptista SD, Vinogradov E, Stajich JE, Filler SG, Howell PL, and Sheppard DC. (2016). Deacetylation of Fungal Exopolysaccharide Mediates Adhesion and Biofilm Formation. mBio. 2016;7(2):e00252-16. DOI:10.1128/mBio.00252-16 |
- Crooks GE, Hon G, Chandonia JM, and Brenner SE. (2004). WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188-90. DOI:10.1101/gr.849004 |