CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Carbohydrate Esterase Family 15"
Harry Brumer (talk | contribs) (Added PDB links) |
Harry Brumer (talk | contribs) |
||
| (3 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
* [[Author]]: ^^^Jenny Arnling Bååth^^^ and ^^^Scott Mazurkewich^^^ | * [[Author]]: ^^^Jenny Arnling Bååth^^^ and ^^^Scott Mazurkewich^^^ | ||
* [[Responsible Curator]]: ^^^Johan Larsbrink^^^ | * [[Responsible Curator]]: ^^^Johan Larsbrink^^^ | ||
| Line 11: | Line 11: | ||
|{{Hl2}} colspan="2" align="center" |'''Carbohydrate Esterase Family CE15''' | |{{Hl2}} colspan="2" align="center" |'''Carbohydrate Esterase Family CE15''' | ||
|- | |- | ||
| − | |''' | + | ||'''Acid/alcohol sugar substrate''' |
| − | | | + | |Acid |
|- | |- | ||
|'''Mechanism''' | |'''Mechanism''' | ||
| − | | | + | |serine hydrolase |
|- | |- | ||
|'''Active site residues''' | |'''Active site residues''' | ||
| − | |known | + | |known, catalytic triad |
|- | |- | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
| Line 27: | Line 27: | ||
<!-- This is the end of the table --> | <!-- This is the end of the table --> | ||
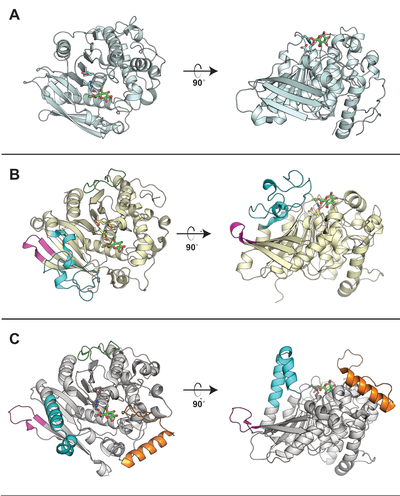
| + | [[File: CE15_CAZypedia_Figure.png|thumb|right|400px|'''Figure 1. Comparison of structurally determined CE15 members.''' The CE15s (A) ''St''GE2 from ''Thermothelomyces thermophila'' (PDB ID [{{PDBlink}}4g4j 4G4J]), (B) ''Ot''CE15A from ''Opitutus terrae'' (PDB ID [{{PDBlink}}6gs0 6GS0]), and (C) ''Tt''CE15A from ''Teredinibacter turnerae'' (PDB ID [{{PDBlink}}6hsw 6HSW]) are shown in cartoon representation. The catalytic triad in each enzyme is shown as sticks and the methyl ester of 4-''O''-methyl glucuronoate first observed in ''St''GE2 is shown in all structures as green sticks. While all CE15 members contain the alpha/beta hydrolase fold, the most prominent difference across the CE15 family observed to-date are the presence, absence, or variety of inserted regions that protrude and build-up ridges around the active site (the differently colored regions in the ''Ot''CE15A and ''Tt''CE15A). The extent to which these regions affect the enzyme’s substrate specificity has yet to be fully elucidated.]] | ||
== Substrate specificity == | == Substrate specificity == | ||
| Line 32: | Line 33: | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | Representative structures of CE15 enzymes from bacterial and fungal sources have been determined, including ''Tr''GE (Cip2) from ''T. reesei'' (''Hypocrea jecorina'' | + | Representative structures of CE15 enzymes from bacterial and fungal sources have been determined, including ''Tr''GE (Cip2) from ''T. reesei'' (''Hypocrea jecorina'', PDB [{{PDBlink}}3pic 3pic]) <cite>Pokkuluri2011</cite>, ''St''GE2 from ''Thermothelomyces thermophila'' (''Sporotrichum thermophile'', PDB [{{PDBlink}}4g4g 4g4g], [{{PDBlink}}4g4i 4g4i], and [{{PDBlink}}4g4j 4g4j]) <cite>Charavgi2013</cite>, marine metagenome sequence MZ0003 (PDB [{{PDBlink}}6ehn 6ehn]) <cite>Desanti2017</cite>, ''Ot''CE15A (PDB [{{PDBlink}}6grw 6grw] and [{{PDBlink}}6gs0 6gs0]) and ''Su''CE15C (PDB [{{PDBlink}}6gry 6gry] and [{{PDBlink}}6gu8 6gu8]) <cite>Arnlingbaath2018</cite> (see the CAZy database for a [http://www.cazy.org/CE15_structure.html continuously updated list]). All structurally determined CE15 enzymes share an alpha/beta hydrolase fold, consisting of a three-layer alpha-beta-alpha sandwich with the active site in a solvent-exposed cleft. The structures of the bacterial enzymes determined thus far exhibit sizeable inserts which result in much deeper active site pockets compared to the shallow active sites seen in fungal glucuronoyl esterase structures <cite>Desanti2017 Arnlingbaath2018 </cite>. |
== Catalytic Residues and Mechanism == | == Catalytic Residues and Mechanism == | ||
Revision as of 09:26, 16 July 2019
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Jenny Arnling Bååth^^^ and ^^^Scott Mazurkewich^^^
- Responsible Curator: ^^^Johan Larsbrink^^^
| Carbohydrate Esterase Family CE15 | |
| Acid/alcohol sugar substrate | Acid |
| Mechanism | serine hydrolase |
| Active site residues | known, catalytic triad |
| CAZy DB link | |
| http://www.cazy.org/CE15.html | |
Substrate specificity
All CE15 enzymes characterized to-date are glucuronoyl esterases, cleaving esters of D-glucuronic acid. The first reported glucuronoyl esterase was ScGE1 from the white-rot fungus Schizophyllum commune, and the activity was demonstrated by TLC on a methyl ester of 4-O-methyl-D-glucuronic acid [1]. While CE15 members are found in both fungal and bacterial species, several bacterial CE15 enzymes are more promiscuous than their fungal counterparts and are active also on esters of galacturonoate [2]. Feruloyl- and acetyl esterase activities have been reported for certain CE15 enzymes as side activities [3, 4]. The proposed physiological role of CE15 enzymes is to hydrolyze lignin-carbohydrate ester linkages between lignin and glucuronoxylan in plant cell walls, and a few studies have demonstrated their activity on lignocellulose-derived materials and plant biomass [4, 5, 6].
Three-dimensional structures
Representative structures of CE15 enzymes from bacterial and fungal sources have been determined, including TrGE (Cip2) from T. reesei (Hypocrea jecorina, PDB 3pic) [7], StGE2 from Thermothelomyces thermophila (Sporotrichum thermophile, PDB 4g4g, 4g4i, and 4g4j) [8], marine metagenome sequence MZ0003 (PDB 6ehn) [9], OtCE15A (PDB 6grw and 6gs0) and SuCE15C (PDB 6gry and 6gu8) [2] (see the CAZy database for a continuously updated list). All structurally determined CE15 enzymes share an alpha/beta hydrolase fold, consisting of a three-layer alpha-beta-alpha sandwich with the active site in a solvent-exposed cleft. The structures of the bacterial enzymes determined thus far exhibit sizeable inserts which result in much deeper active site pockets compared to the shallow active sites seen in fungal glucuronoyl esterase structures [2, 9].
Catalytic Residues and Mechanism
All CE15 enzymes are serine-type hydrolases, containing a catalytic triad of Glu/Asp-His-Ser [2, 7, 8, 9]. The position of the acidic residue of the triad is not similarly positioned in all CE15 members as the residue can be found on different loops of the conserved fold [9]. A conserved arginine found in all of the CE15 structures, proximal to the catalytic triad, has been proposed to stabilize the formation of the oxyanion during catalysis [2].
Family Firsts
- First 3-D structure
- The first solved structure of a CE15 enzyme was the Cip2 catalytic domain from Trichoderma reesei (TrGE) [7].
- First mechanistic insight
- The crystal structure of StGE2 (from Sporotrichum thermophile) in complex with the ligand 4-O-methyl-beta-D-glucopyranuronate gave the first direct insight into substrate binding [8].
References
- Spániková S and Biely P. (2006). Glucuronoyl esterase--novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett. 2006;580(19):4597-601. DOI:10.1016/j.febslet.2006.07.033 |
- Arnling Bååth J, Mazurkewich S, Knudsen RM, Poulsen JN, Olsson L, Lo Leggio L, and Larsbrink J. (2018). Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion. Biotechnol Biofuels. 2018;11:213. DOI:10.1186/s13068-018-1213-x |
- De Santi C, Willassen NP, and Williamson A. (2016). Biochemical Characterization of a Family 15 Carbohydrate Esterase from a Bacterial Marine Arctic Metagenome. PLoS One. 2016;11(7):e0159345. DOI:10.1371/journal.pone.0159345 |
- Mosbech C, Holck J, Meyer AS, and Agger JW. (2018). The natural catalytic function of CuGE glucuronoyl esterase in hydrolysis of genuine lignin-carbohydrate complexes from birch. Biotechnol Biofuels. 2018;11:71. DOI:10.1186/s13068-018-1075-2 |
- d'Errico C, Börjesson J, Ding H, Krogh KB, Spodsberg N, Madsen R, and Monrad RN. (2016). Improved biomass degradation using fungal glucuronoyl-esterases-hydrolysis of natural corn fiber substrate. J Biotechnol. 2016;219:117-23. DOI:10.1016/j.jbiotec.2015.12.024 |
- Arnling Bååth J, Giummarella N, Klaubauf S, Lawoko M, and Olsson L. (2016). A glucuronoyl esterase from Acremonium alcalophilum cleaves native lignin-carbohydrate ester bonds. FEBS Lett. 2016;590(16):2611-8. DOI:10.1002/1873-3468.12290 |
- Pokkuluri PR, Duke NE, Wood SJ, Cotta MA, Li XL, Biely P, and Schiffer M. (2011). Structure of the catalytic domain of glucuronoyl esterase Cip2 from Hypocrea jecorina. Proteins. 2011;79(8):2588-92. DOI:10.1002/prot.23088 |
- Charavgi MD, Dimarogona M, Topakas E, Christakopoulos P, and Chrysina ED. (2013). The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 1):63-73. DOI:10.1107/S0907444912042400 |
- De Santi C, Gani OA, Helland R, and Williamson A. (2017). Structural insight into a CE15 esterase from the marine bacterial metagenome. Sci Rep. 2017;7(1):17278. DOI:10.1038/s41598-017-17677-4 |