CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 104"
| Line 29: | Line 29: | ||
== Substrate specificities == | == Substrate specificities == | ||
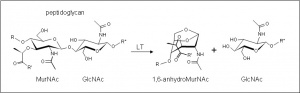
| − | [[Image:LTreaction.jpg|thumb|right|'''Figure 1.''' Reaction catalyzed by family | + | [[Image:LTreaction.jpg|thumb|right|'''Figure 1.''' Reaction catalyzed by family GH104 enzymes. LT, lytic transglycosylase. (''click to enlarge'').]]The glycoside hydrolases of this family are lytic transglyosylases (also referred to as peptidoglycan lyases) of bacteriophage origin and they constitute family 4 of the classification scheme of Blackburn and Clarke <cite>1</cite>. Unfortunately, the origin of many of the hypothetical enzymes listed in GH104 is misleading because they are encoded within prophages which have been integrated into the chromosome of their bacterial host. In other cases, the phage enzyme has been acquired by pathogenicity islands on the bacterial chromosome. The prototype for this family is the enzyme from ''lambda'' phage. |
These enzymes cleave the β-1,4 linkage between ''N''-acetylmuramoyl and ''N''-acetylglucosaminyl residues in peptidoglycan (Figure 1). No other activities have been observed. | These enzymes cleave the β-1,4 linkage between ''N''-acetylmuramoyl and ''N''-acetylglucosaminyl residues in peptidoglycan (Figure 1). No other activities have been observed. | ||
| Line 39: | Line 39: | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | As with other lytic transglycosylases (families [[GH23]], [[GH102]], and [[GH103]]), the GH104 enzymes are thought to possess a single catalytic acid/base residue. This residue in | + | As with other lytic transglycosylases (families [[GH23]], [[GH102]], and [[GH103]]), the GH104 enzymes are thought to possess a single catalytic acid/base residue. This residue in lambda bacteriophage lytic transglycosylase has been inferred by X-ray crystallography as Glu19 <cite>3</cite>. The mechanism of action of the family GH104 enzymes has not been investigated and thus it is not known if they follow that of the lytic transglycosylases of families [[GH23]], [[GH102]], or [[GH103]]. |
| Line 47: | Line 47: | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First identification of lytic transglycosylase: Bacteriophage | + | ;First identification of lytic transglycosylase: Bacteriophage lambda <cite>3</cite>. |
;First catalytic nucleophile identification: Not applicable for lytic transglycosylases. | ;First catalytic nucleophile identification: Not applicable for lytic transglycosylases. | ||
| − | ;First general acid/base residue identification: Inferred by X-ray crystallography of | + | ;First general acid/base residue identification: Inferred by X-ray crystallography of lambda bacteriophage <cite>3</cite>. |
| − | ;First 3-D structure: | + | ;First 3-D structure: Lambda bacteriophage <cite>3</cite>. |
| Line 57: | Line 57: | ||
#1 pmid=11139297 | #1 pmid=11139297 | ||
#2 pmid=357 | #2 pmid=357 | ||
| − | #3 pmid=11341831 | + | #3 pmid=9514719 |
| + | #4 pmid=11341831 | ||
</biblio> | </biblio> | ||
Revision as of 22:11, 20 February 2010
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Anthony Clarke^^^
- Responsible Curator: ^^^Anthony Clarke^^^
| Glycoside Hydrolase Family GHnn | |
| Clan | none |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/fam/GHnn.html | |
Substrate specificities
The glycoside hydrolases of this family are lytic transglyosylases (also referred to as peptidoglycan lyases) of bacteriophage origin and they constitute family 4 of the classification scheme of Blackburn and Clarke [1]. Unfortunately, the origin of many of the hypothetical enzymes listed in GH104 is misleading because they are encoded within prophages which have been integrated into the chromosome of their bacterial host. In other cases, the phage enzyme has been acquired by pathogenicity islands on the bacterial chromosome. The prototype for this family is the enzyme from lambda phage.
These enzymes cleave the β-1,4 linkage between N-acetylmuramoyl and N-acetylglucosaminyl residues in peptidoglycan (Figure 1). No other activities have been observed.
Kinetics and Mechanism
The lytic transglycosidases, strictly speaking, are retaining enzymes. However, they are not hydrolases but rather catalyse an intramolecular glycosyl transferase reaction onto the C-6 hydroxyl group of the muramoyl residue leading to the generation of a terminal 1,6-anhdyromuramoyl product (Figure 1) thus lacking a reducing end [2]. No detailed analyses involving either steady state or pre-steady state kinetic studies have been reported.
Catalytic Residues
As with other lytic transglycosylases (families GH23, GH102, and GH103), the GH104 enzymes are thought to possess a single catalytic acid/base residue. This residue in lambda bacteriophage lytic transglycosylase has been inferred by X-ray crystallography as Glu19 [3]. The mechanism of action of the family GH104 enzymes has not been investigated and thus it is not known if they follow that of the lytic transglycosylases of families GH23, GH102, or GH103.
Three-dimensional structures
The three-dimensional structure of only the lambda phage enzyme has been solved [4], and like the other lytic transglycosylases of families GH23, and GH103, it possesses the well characterized α+β “lysozyme fold.”
Family Firsts
- First identification of lytic transglycosylase
- Bacteriophage lambda [3].
- First catalytic nucleophile identification
- Not applicable for lytic transglycosylases.
- First general acid/base residue identification
- Inferred by X-ray crystallography of lambda bacteriophage [3].
- First 3-D structure
- Lambda bacteriophage [3].
References
- Blackburn NT and Clarke AJ. (2001). Identification of four families of peptidoglycan lytic transglycosylases. J Mol Evol. 2001;52(1):78-84. DOI:10.1007/s002390010136 |
- Höltje JV, Mirelman D, Sharon N, and Schwarz U. (1975). Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124(3):1067-76. DOI:10.1128/jb.124.3.1067-1076.1975 |
- Evrard C, Fastrez J, and Declercq JP. (1998). Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes. J Mol Biol. 1998;276(1):151-64. DOI:10.1006/jmbi.1997.1499 |
- Leung AK, Duewel HS, Honek JF, and Berghuis AM. (2001). Crystal structure of the lytic transglycosylase from bacteriophage lambda in complex with hexa-N-acetylchitohexaose. Biochemistry. 2001;40(19):5665-73. DOI:10.1021/bi0028035 |