CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 106"
| Line 35: | Line 35: | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | Structural characterization of ''B. thetaiotaomicron'' BT0986 identified two highly conserved carboxylate amino acids (E461 and E593) that were essential for activity. These glutamates are separated by 8.0 Å, a distance between catalytic residues consistent with an inverting mechanism. Additionally, E593, which is 5.8 Å from the anomeric carbon is ideally positioned to act as general base and E461 is the general acid <cite>Ndeh2017</cite>. | + | [[File:BT0986_2.png|thumb|300px|right|'''Figure 1.''' '''BT0986 active site.''' The catalytic amino acids are shown in orange and yellow are the remaining key residues present in the active site. The calcium ion in the active site is shown as a cyan sphere and its polar contacts indicated by black dashed lines. D-rhamnopyranose tetrazole is represented in green.]] |
| + | Structural characterization of ''B. thetaiotaomicron'' BT0986 identified two highly conserved carboxylate amino acids (E461 and E593) that were essential for activity (Figure 1). These glutamates are separated by 8.0 Å, a distance between catalytic residues consistent with an inverting mechanism. Additionally, E593, which is 5.8 Å from the anomeric carbon is ideally positioned to act as general base and E461 is the general acid <cite>Ndeh2017</cite>. | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The three-dimensional structure of ''B. thetaiotaomicron'' BT0986 solved using X-ray crystallography represents the first structure of an GH106 enzyme ([https://www.rcsb.org/structure/5MQN 5MQN]). BT0986 displays a N-terminal catalytic domain that presents an ( | + | [[File:BT0986_1.png|thumb|300px|right|'''Figure 2.''' '''BT0986 structure'''([https://www.rcsb.org/structure/5MQN 5MQN]). The figure shows the (β/α)<sub>8</sub>-barrel domain (1, yellow) and the additional two b-stranded domains (2 to 6). The D-rhamnopyranose tetrazole represented as sticks with carbon in green and the calcium ion (cyan sphere) are shown at the centre of the (β/α)<sub>8</sub>-barrel.]] |
| + | The three-dimensional structure of ''B. thetaiotaomicron'' BT0986 solved using X-ray crystallography represents the first structure of an GH106 enzyme ([https://www.rcsb.org/structure/5MQN 5MQN]). BT0986 displays a N-terminal catalytic domain that presents an (β/α)<sub>8</sub>-barrel fold with several appended several β-stranded domains (C-terminal) (Figure 2) <cite>Ndeh2017</cite>. | ||
== Family Firsts == | == Family Firsts == | ||
Revision as of 16:13, 11 February 2018
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Ana Luis^^^
- Responsible Curator: ^^^Harry Gilbert^^^
| Glycoside Hydrolase Family GH106 | |
| Clan | none |
| Mechanism | inverting |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH106.html | |
Substrate specificities
The glycoside hydrolases of this family are alfa L-rhamnosidases (EC 3.2.1.40). The first GH106 characterized was Rham from Sphingomonas paucimobilis FP2001. This enzyme showed activity against p-nitrophenyl α-L-rhamnopyranoside [1]. More recently, two Bacteroides thetaiotaomicron enzymes (BT0986 and BT4145) have been characterized. These enzymes are exo active against linkages present in pectin polysaccharides. BT0986 cleaves the L-Rha-α-1,2-L-Arap linkage in the terminal region of Chain B of rhamnogalacturonan II [2]. The enzyme BT4145 targets the L-Rha-α-1,4-D-GalA linkage in the backbone of rhamnogalacturonan I [3]. All of genes encoding family 106 members are found in bacteria (CAZy).
Kinetics and Mechanism
Family GH106 members act by inverting the anomeric configuration of the glycone sugar participating in the scissle glycosidic linkage (inverting mechanism), and thus mediate bond cleave through a single displacement mechanism [3]. Additionally, GH106 enzymes are Ca 2+ dependent [2]. The only other ion dependent enzyme families are the exo-α-mannosidases GH38, GH47 and GH92. The structure of BT0986 in complex with D-rhamnopyranose tetrazole indicates that catalysis is proceeds via a B2,5 transition state [2].
Catalytic Residues
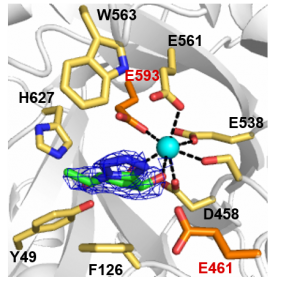
Structural characterization of B. thetaiotaomicron BT0986 identified two highly conserved carboxylate amino acids (E461 and E593) that were essential for activity (Figure 1). These glutamates are separated by 8.0 Å, a distance between catalytic residues consistent with an inverting mechanism. Additionally, E593, which is 5.8 Å from the anomeric carbon is ideally positioned to act as general base and E461 is the general acid [2].
Three-dimensional structures
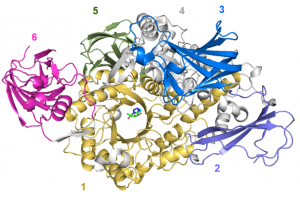
The three-dimensional structure of B. thetaiotaomicron BT0986 solved using X-ray crystallography represents the first structure of an GH106 enzyme (5MQN). BT0986 displays a N-terminal catalytic domain that presents an (β/α)8-barrel fold with several appended several β-stranded domains (C-terminal) (Figure 2) [2].
Family Firsts
- First stereochemistry determination
- BT4145 from Bacteroides thetaiotaomicron [3].
- First catalytic nucleophile identification
- BT0986 from Bacteroides thetaiotaomicron [2].
- First general acid/base residue identification
- BT0986 from Bacteroides thetaiotaomicron [2].
- First 3-D structure
- BT0986 from Bacteroides thetaiotaomicron [2].
References
-
Miyata T, Kashige N, Satho T, Yamaguchi T, Aso Y and Miake F.Cloning (2005) Sequence analysis, and expression of the gene encoding Sphingomonas paucimobilis FP2001 alpha-L-rhamnosidase. Curr Microbiol, vol 51, no. 2., pp. 105-109.
- Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O'Neil MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet MC, Martens EC, Henrissat B, and Gilbert HJ. (2017). Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544(7648):65-70. DOI:10.1038/nature21725 |
- Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet MC, Henrissat B, Martens EC, Mosimann SC, Abbott DW, and Gilbert HJ. (2018). Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol. 2018;3(2):210-219. DOI:10.1038/s41564-017-0079-1 |