CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Glycoside Hydrolase Family 107
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^David Teze^^^
- Responsible Curator: ^^^Al Boraston^^^
| Glycoside Hydrolase Family GH107 | |
| Clan | GH-R |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH107.html | |
Substrate specificities
The currently characterized glycoside hydrolases of this family are endo-acting α-fucosidases active on sulfated fucans (or fucoidans) from brown algae. All described GH107 family members are endo-1,4-fucanase of bacterial origin, and together with enzymes from the CAZY family GH29, they form the clan GH-R. Sequences of GH107 family members were first reported in 2006,[1] even though the enzymatic activity was reported earlier.
Kinetics and Mechanism
The mechanism was proven to be retaining by observation of the formation of an α-O-linked mercaptoethanol on a L-Fuc-2,3-disulfate-(α1-3)-L-Fuc-2-sulfate disaccharide by transglycosylation.[2] It should then follow a classical Koshland double-displacement mechanism similarly to GH29.
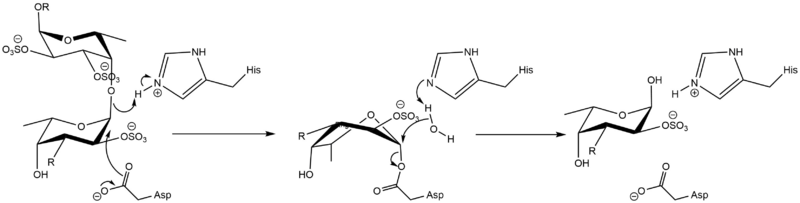
Catalytic Residues
The catalytic nucleophile is an aspartate, while the catalytic acid-base is a histidine. The later is unusual in GHs, and a divergence from GH29, but is likely necessary to avoid electronic repulsion with the substrate sulfate groups. These two residues have been identified by structural superimposition with GH29 enzymes, and are conserved within the few members of the GH107 family. The catalytic His has been further confirmed by the lack of activity of the H294Q mutant of Mariniflexile fucanivorans.[2] The catalytic aspartate was also proposed to be one of the catalytic residue on sequence analysis alone, in a simultaneously released paper.[3]
Three-dimensional structures
The crystal structures of Mariniflexile fucanivorans (PDB: 6dns,6dms,6dlh) and Psychromonas sp. (PDB: 6m8n) have been determined in 2018.[2] The Psychromonas sp. (PDB: 6m8n) enzyme showed a single catalytic domain with a (β/α)8 / TIM-barrel fold, while in the Mariniflexile fucanivorans enzyme, this catalytic domain is followed by three Ig-like domains that wrap around the catalytic one.[2]
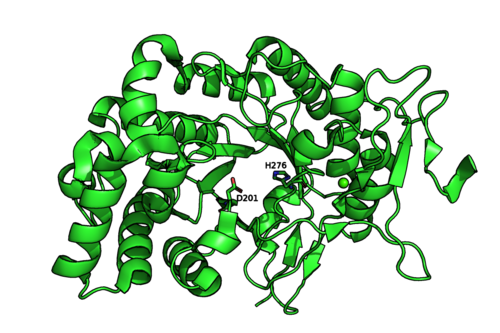
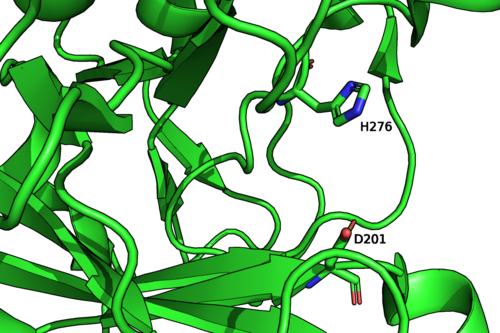
Figure 2: Psychromonas sp. GH107 structure and catalytic residues (PDB: 6m8n).
Family Firsts
- First stereochemistry determination
- The retaining mechanism was determined in 2018.[2]
- First catalytic nucleophile identification
- An aspartic acid side chain acting as a catalytic nucleophile was identified by two simultaneous studies in the Autumn of 2018.[2][3]
- First general acid/base residue identification
- A histidine sidechain acting as a catalytic acid-base residue was identified in 2018.[2]
- First 3-D structure
- The crystal structures of Mariniflexile fucanivorans (PDB: 6dns,6dms,6dlh) and Psychromonas sp. (PDB: 6m8n) have been released at the same time, in 2018.[2]
References
- Colin S, Deniaud E, Jam M, Descamps V, Chevolot Y, Kervarec N, Yvin JC, Barbeyron T, Michel G, and Kloareg B. (2006). Cloning and biochemical characterization of the fucanase FcnA: definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology. 2006;16(11):1021-32. DOI:10.1093/glycob/cwl029 |
- Vickers C, Liu F, Abe K, Salama-Alber O, Jenkins M, Springate CMK, Burke JE, Withers SG, and Boraston AB. (2018). Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to α-l-fucosidases from GH29. J Biol Chem. 2018;293(47):18296-18308. DOI:10.1074/jbc.RA118.005134 |
- Schultz-Johansen M, Cueff M, Hardouin K, Jam M, Larocque R, Glaring MA, Hervé C, Czjzek M, and Stougaard P. (2018). Discovery and screening of novel metagenome-derived GH107 enzymes targeting sulfated fucans from brown algae. FEBS J. 2018;285(22):4281-4295. DOI:10.1111/febs.14662 |