CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Glycoside Hydrolase Family 11
| Glycoside Hydrolase Family GH11 | |
| Clan | GH-C |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/fam/GH11.html | |
Substrate specificities
The enzymes in this family are endo-β-1,4-xylanases. No other activities have been observed.
Kinetics and Mechanism
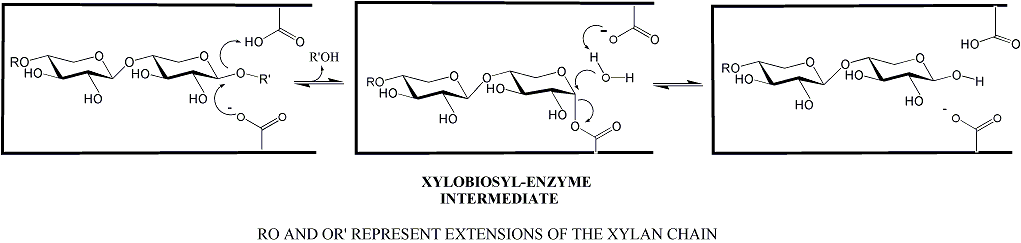
Family GH11 xylanases are retaining enzymes, as first shown by NMR [1] and follow a classical Koshland double-displacement mechanism. No detailed analyses involving both steady state and pre-steady state kinetic studies have been reported. However, recent studies of the contributions of each substrate hydroxyl to transition state binding, thus to catalysis, have been reported [2].
Catalytic Residues
The catalytic nucleophile was first identified in the Bacillus circulans endo-xylanase as Glu78 in the sequence ITELD through trapping of the 2-deoxy-2-fluoroxylobiosyl-enzyme intermediate and subsequent peptide mapping via LC-MS/MS technologies [3]. The importance of the precise positioning of the nucleophile was probed by detailed kinetic analysis of mutants modified at that position [4]. The acid/base catalyst was first identified as Glu127 in this same enzyme through detailed mechanistic analysis of mutants at that position, which included azide rescue experiments [5]. Indeed detailed analyses of the role of the acid/base catalyst have been described. NMR based pKa measurements through the catalytic cycle reveal pKa cycling [6] while consequences of repositioning and removing the acid/base residue reveal its contributions [7].
Three-dimensional structures
Three-dimensional structures are available for a number of Family GH11 enzymes, the first solved being that of the Bacillus circulans xylanase [8]. As members of Clan GH-C they have a jellyroll fold. NMR assignment has allowed insights into both dynamics and electrostatics within the protein [8], [9], [10], [11]. .
Family Firsts
- First sterochemistry determination
- Bacillus circulans endo-xylanase Bcx by NMR [1]
- First nuc. ID
- Bacillus circulans endo-xylanase Bcx by 2-fluoroglucose labeling [3]
- First A/B ID
- Bacillus circulans fimi endo-xylanase Bcx by rescue kinetics with mutants [5]
- First 3-D structures of a (major subfamily)
- Bacillus circulans endo-xylanase Bcx [8]
References
- Gebler J, Gilkes NR, Claeyssens M, Wilson DB, Béguin P, Wakarchuk WW, Kilburn DG, Miller RC Jr, Warren RA, and Withers SG. (1992). Stereoselective hydrolysis catalyzed by related beta-1,4-glucanases and beta-1,4-xylanases. J Biol Chem. 1992;267(18):12559-61. | Google Books | Open Library
-
pmid=Wicki In Press
- Miao S, Ziser L, Aebersold R, and Withers SG. (1994). Identification of glutamic acid 78 as the active site nucleophile in Bacillus subtilis xylanase using electrospray tandem mass spectrometry. Biochemistry. 1994;33(23):7027-32. DOI:10.1021/bi00189a002 |
- Lawson SL, Wakarchuk WW, and Withers SG. (1996). Effects of both shortening and lengthening the active site nucleophile of Bacillus circulans xylanase on catalytic activity. Biochemistry. 1996;35(31):10110-8. DOI:10.1021/bi960586v |
- MacLeod AM, Lindhorst T, Withers SG, and Warren RA. (1994). The acid/base catalyst in the exoglucanase/xylanase from Cellulomonas fimi is glutamic acid 127: evidence from detailed kinetic studies of mutants. Biochemistry. 1994;33(20):6371-6. DOI:10.1021/bi00186a042 |
- McIntosh LP, Hand G, Johnson PE, Joshi MD, Körner M, Plesniak LA, Ziser L, Wakarchuk WW, and Withers SG. (1996). The pKa of the general acid/base carboxyl group of a glycosidase cycles during catalysis: a 13C-NMR study of bacillus circulans xylanase. Biochemistry. 1996;35(31):9958-66. DOI:10.1021/bi9613234 |
- Wakarchuk WW, Campbell RL, Sung WL, Davoodi J, and Yaguchi M. (1994). Mutational and crystallographic analyses of the active site residues of the Bacillus circulans xylanase. Protein Sci. 1994;3(3):467-75. DOI:10.1002/pro.5560030312 |
- Plesniak LA, Wakarchuk WW, and McIntosh LP. (1996). Secondary structure and NMR assignments of Bacillus circulans xylanase. Protein Sci. 1996;5(6):1118-35. DOI:10.1002/pro.5560050614 |
- Joshi MD, Hedberg A, and McIntosh LP. (1997). Complete measurement of the pKa values of the carboxyl and imidazole groups in Bacillus circulans xylanase. Protein Sci. 1997;6(12):2667-70. DOI:10.1002/pro.5560061224 |
- Connelly GP, Withers SG, and McIntosh LP. (2000). Analysis of the dynamic properties of Bacillus circulans xylanase upon formation of a covalent glycosyl-enzyme intermediate. Protein Sci. 2000;9(3):512-24. DOI:10.1110/ps.9.3.512 |
- Lawson SL, Wakarchuk WW, and Withers SG. (1997). Positioning the acid/base catalyst in a glycosidase: studies with Bacillus circulans xylanase. Biochemistry. 1997;36(8):2257-65. DOI:10.1021/bi9620215 |