CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Glycoside Hydrolase Family 121
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Kiyotaka Fujita^^^
- Responsible Curator: ^^^Shinya Fushinobu^^^
| Glycoside Hydrolase Family GH121 | |
| Clan | none |
| Mechanism | retaining |
| Active site residues | not known |
| CAZy DB link | |
| http://www.cazy.org/GH121.html | |
Substrate specificities
This family of glycoside hydrolases contains β-L-arabinobiosidases, as demonstrated for HypBA2 from Bifidobacterium longum JCM 1217 [1]. HypBA2 liberates the disaccharide Arafβ1-2Araf (β-Ara2, a substrate of the GH127 β-L-arabinofuranosidase from B. longum JCM 1217 [2]) from unmodified Arafβ1-2Arafβ1-2Arafβ-hydroxyproline (Ara3-Hyp), but not Arafα1-3Arafβ1-2Arafβ1-2Arafβ-Hyp (Ara4-Hyp) or Arafβ1-2Arafβ-Hyp (Ara2-Hyp). HypBA2 directly liberates β-Ara2 from hydroxyproline-rich glycoproteins (HRGPs) such as carrot extensin and potato lectin. The family members are only found from prokaryote genomes, such as bacteria and actinomycetes.
Kinetics and Mechanism
HypBA2 is a retaining enzyme. The stereochemical course of the reaction was shown by transglycosylation activity toward 1-alkanols, such as methanol; the resulting Arafβ1-2Arafβ-Me was identified by 1H-NMR and 13C-NMR analysis.
Catalytic Residues
The catalytic residues are not known but three conserved residues (Glu373, Asp515, and Glu713 in B. longum HypBA2) are the candidates based on mutagenesis and structural comparison [3].
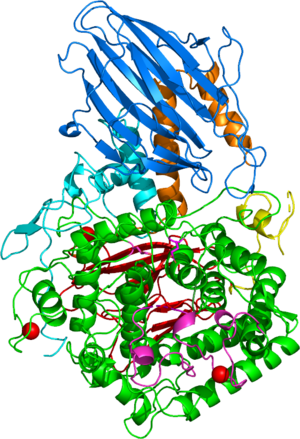
Three-dimensional structures
The first solved 3-D structure was β-L-arabinobiosidase HypBA2 from Bifidobacterium longum (PDB 6M5A) [3]. The catalytic domain adops an (α/α)6 barrel fold similar to GH142, GH63, GH78, GH94, and GH37 (Figure 1).
Family Firsts
- First stereochemistry determination
- Shown to be retaining for HypBA2 enzyme by measurement of glycosyl transfer reactions to methanol and the 1H-NMR and 13C-NMR spectra [1].
- First catalytic nucleophile identification
- Predicted based on structural homology [3], but currently no experimental proof.
- First general acid/base residue identification
- Predicted based on structural homology [3], but currently no experimental proof.
- First 3-D structure
- β-L-arabinobiosidase HypBA2 from Bifidobacterium longum [3].
References
- Fujita K, Sakamoto S, Ono Y, Wakao M, Suda Y, Kitahara K, and Suganuma T. (2011). Molecular cloning and characterization of a beta-L-Arabinobiosidase in Bifidobacterium longum that belongs to a novel glycoside hydrolase family. J Biol Chem. 2011;286(7):5143-50. DOI:10.1074/jbc.M110.190512 |
- Fujita K, Takashi Y, Obuchi E, Kitahara K, and Suganuma T. (2011). Characterization of a novel β-L-Arabinofuranosidase in Bifidobacterium longum: functional elucidation of A DUF1680 family member. J Biol Chem. 2011;286(44):38079-38085. DOI:10.1074/jbc.M111.248690 |
- Saito K, Viborg AH, Sakamoto S, Arakawa T, Yamada C, Fujita K, and Fushinobu S. (2020). Crystal structure of β-L-arabinobiosidase belonging to glycoside hydrolase family 121. PLoS One. 2020;15(6):e0231513. DOI:10.1371/journal.pone.0231513 |