CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 141"
| Line 39: | Line 39: | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
[[File:BT1002.png|thumb|300px|right|'''Figure 1.''' '''BT1002 strucutre'''. ([https://www.rcsb.org/structure/5MQP PDB ID 5PDB]) The figure shows the β-parallel catalytic domain (purple) and the additional β-sandwich domain (yellow)]] | [[File:BT1002.png|thumb|300px|right|'''Figure 1.''' '''BT1002 strucutre'''. ([https://www.rcsb.org/structure/5MQP PDB ID 5PDB]) The figure shows the β-parallel catalytic domain (purple) and the additional β-sandwich domain (yellow)]] | ||
| − | The three-dimensional structure has been solved for ''B. thetaiotaomicron'' BT1002 at 2 Å ([https://www.rcsb.org/structure/5MQP PDB ID 5PDB]). The protein has two domains: a N-terminal β-sandwich and a C-terminal catalytic domain that folds into a right-handed parallel β-helix . The overall fold displays similarity to members of polysaccharide lyase family 1 ([[PL1]]) and glycoside hydrolase family 28 ([[GH28]]). (Figure 1) <cite>Ndeh2017</cite>. The active site pocket is in a similar location to the catalytic centre of GH120 β-xylosidases <cite></cite> | + | |
| + | The three-dimensional structure has been solved for ''B. thetaiotaomicron'' BT1002 at 2 Å ([https://www.rcsb.org/structure/5MQP PDB ID 5PDB]). The protein has two domains: a N-terminal β-sandwich and a C-terminal catalytic domain that folds into a right-handed parallel β-helix . The overall fold displays similarity to members of polysaccharide lyase family 1 ([[PL1]]) and glycoside hydrolase family 28 ([[GH28]]). (Figure 1) <cite>Ndeh2017</cite>. The active site pocket is in a similar location to the catalytic centre of GH120 β-xylosidases <cite>Huang2012</cite>. The catalytic resisdues identified in the GH120 β-xylosidase, however, are not conserved in the Gh141 α-L-fucosidase talytic residues identified in a GH120 in are not conserved in | ||
== Family Firsts == | == Family Firsts == | ||
| Line 51: | Line 52: | ||
#Ndeh2017 pmid=28329766 | #Ndeh2017 pmid=28329766 | ||
#Heinze2017 pmid=28894250 | #Heinze2017 pmid=28894250 | ||
| + | |||
| + | #Huang2012 pmid=22992047 | ||
</biblio> | </biblio> | ||
[[Category:Glycoside Hydrolase Families|GH141]] | [[Category:Glycoside Hydrolase Families|GH141]] | ||
Revision as of 11:25, 13 February 2018
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Ana Luis^^^
- Responsible Curator: ^^^Harry Gilbert^^^
| Glycoside Hydrolase Family GH141 | |
| Clan | none |
| Mechanism | unknown |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH141.html | |
Substrate specificities
Glycoside hydrolases of family 141 (CAZy) display α-L-fucosidase (EC 3.2.1.51) [1] or xylanase (EC 3.2.1.8) [2]activities. The Bacteroides thetaiotaomicron enzyme BT1002 was the founding member of this family. The enzyme cleaves the fucosodic linkage in 2-O-methyl-D-xylose-α-1,3-L-fucose-α1,4-L-Rhap, a component of Chain B of rhamnogalacturonan II, a complex pectin conserved in the primary cell walls [1]. Recently, an endo-xylanase from Clostridium thermocellum (Xyn141E) was also described. Xyn141E is active against a range of xylans, displaying lowl elvel side activity against other β1,4-glycans such as carboxymethyl cellulose, barley beta glucan and mannan from ivory nut [2].
Kinetics and Mechanism
Very little is known about the kinetics or mechanism of GH141 enzymes. However, in the BT1002 crystal structure, the distance of 6.1 Å between the catalytic residues suggests that members of this family may be retaining enzymes and follow a double displacement mechanism [1].
Catalytic Residues
In the BT1002 crystal structure, two aspartates (Asp523 and Asp564), which are highly conserved and located within the active site pocket, are the proposed catalytic residues. Site directed mutagenesis of these amino acids abolished the catalytic activity of BT1002 indicating the essential role in catalysis. Additionally, the structural location of the catalytic residues suggests that Asp523 (at the base of the pocket) acts as catalytic nucleophile and Asp564 (at the lip of the active site) is the general acid-base residue [1].
Three-dimensional structures
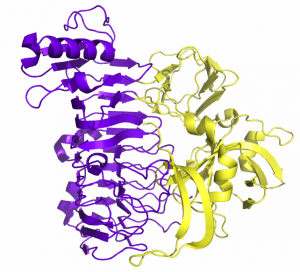
The three-dimensional structure has been solved for B. thetaiotaomicron BT1002 at 2 Å (PDB ID 5PDB). The protein has two domains: a N-terminal β-sandwich and a C-terminal catalytic domain that folds into a right-handed parallel β-helix . The overall fold displays similarity to members of polysaccharide lyase family 1 (PL1) and glycoside hydrolase family 28 (GH28). (Figure 1) [1]. The active site pocket is in a similar location to the catalytic centre of GH120 β-xylosidases [3]. The catalytic resisdues identified in the GH120 β-xylosidase, however, are not conserved in the Gh141 α-L-fucosidase talytic residues identified in a GH120 in are not conserved in
Family Firsts
- First stereochemistry determination
- Currently unknown.
- First catalytic nucleophile identification
- BT1002 from Bacteroides thetaiotaomicron [1].
- First general acid/base residue identification
- BT1002 from Bacteroides thetaiotaomicron [1].
- First 3-D structure
- BT1002 from Bacteroides thetaiotaomicron [1].
References
- Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O'Neil MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet MC, Martens EC, Henrissat B, and Gilbert HJ. (2017). Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544(7648):65-70. DOI:10.1038/nature21725 |
- Heinze S, Mechelke M, Kornberger P, Liebl W, Schwarz WH, and Zverlov VV. (2017). Identification of endoxylanase XynE from Clostridium thermocellum as the first xylanase of glycoside hydrolase family GH141. Sci Rep. 2017;7(1):11178. DOI:10.1038/s41598-017-11598-y |
- Huang CH, Sun Y, Ko TP, Chen CC, Zheng Y, Chan HC, Pang X, Wiegel J, Shao W, and Guo RT. (2012). The substrate/product-binding modes of a novel GH120 β-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485. Biochem J. 2012;448(3):401-7. DOI:10.1042/BJ20121359 |