CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 174"
Guanchen Liu (talk | contribs) |
Guanchen Liu (talk | contribs) |
||
| Line 31: | Line 31: | ||
== Substrate specificities == | == Substrate specificities == | ||
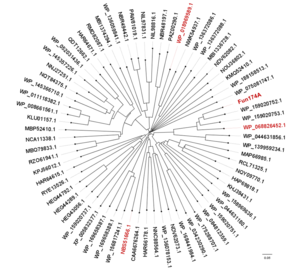
| − | Members of [[GH174|glycoside hydrolase family 174]] have been shown to exhibit α-1,3-L-fucanase activity. The first member of this family, Fun174A from a marine bacterium ''Wenyingzhuangia aestuarii'' OF219, specifically hydrolyze the α-1,3- L-fucoside bonds between 2-O-sulfated and non-sulfated fucose residues in the sulfated fucan from sea cucumber ''Isostichopus badionotus'' in a processive endo-acting manner.< | + | Members of [[GH174|glycoside hydrolase family 174]] have been shown to exhibit α-1,3-L-fucanase activity. The first member of this family, Fun174A from a marine bacterium ''Wenyingzhuangia aestuarii'' OF219, specifically hydrolyze the α-1,3- L-fucoside bonds between 2-O-sulfated and non-sulfated fucose residues in the sulfated fucan from sea cucumber ''Isostichopus badionotus'' in a processive endo-acting manner.<cite>Liu2023</cite> Meanwhile, three homologs of Fun174A from ''Rubritalea marina'', ''Spartobacteria bacterium'' and ''Wenyingzhuangia fucanilytica'', display activities toward sulfated fucan from ''Isostichopus badionotus''.<cite>Liu2023</cite> All 92 members (up to May,2023) of GH174 are bacterial enzymes. |
| − | [[File:Tree.png|thumb]] | + | [[File:Tree.png|thumb|'''Figure 1. The phylogenetic tree of GH174 homologs.''' Sequences comfirmed to exhibit α-1,3-L-fucanase activity were in red.]] |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | The catalytic mechanism of GH174 has not been identified. As mentioned in the report, Fun174A showed no transglycosylating activity in the tested acceptor substrates, such as glycerin, methanol, L-fucose, D-glucose, D-galactose, D-fructose, D-mannose, D-glucosamine and N-acetyl- D-glucosamine.< | + | The catalytic mechanism of GH174 has not been identified. As mentioned in the report, Fun174A showed no transglycosylating activity in the tested acceptor substrates, such as glycerin, methanol, L-fucose, D-glucose, D-galactose, D-fructose, D-mannose, D-glucosamine and N-acetyl- D-glucosamine.<cite>Liu2023</cite> |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | Multiple sequence alignments of GH174 homologs showed that D119, E120 and E218 in Fun174A were highly conserved in all sequences. Three single-site mutants D119E, E120A and E218Q were established, expressed and identified in the report. Mutant D119E, E120A and E218Q resulted in 100.0%, 85.7% and 88.3% loss of activity on sulfated fucan from ''Isostichopus badionotus'' respectively. It indicated that D119, E120 and E218 were critical for the functioning of Fun174A.< | + | Multiple sequence alignments of GH174 homologs showed that D119, E120 and E218 in Fun174A were highly conserved in all sequences. Three single-site mutants D119E, E120A and E218Q were established, expressed and identified in the report. Mutant D119E, E120A and E218Q resulted in 100.0%, 85.7% and 88.3% loss of activity on sulfated fucan from ''Isostichopus badionotus'' respectively. It indicated that D119, E120 and E218 were critical for the functioning of Fun174A.<cite>Liu2023</cite> |
| − | [[File:Weblogo.jpg|thumb]] | + | [[File:Weblogo.jpg|thumb|'''Figure 2. Multiple sequence alignments of residues in GH174 homologs.''' The highly conserved acidic amino acids in all sequences were indicated with black triangles.]] |
== Three-dimensional structures == | == Three-dimensional structures == | ||
No three-dimensional structure has been solved in this glycoside hydrolase family at present. | No three-dimensional structure has been solved in this glycoside hydrolase family at present. | ||
Revision as of 18:42, 25 May 2023
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| Glycoside Hydrolase Family GH174 | |
| Clan | GH-x |
| Mechanism | retaining/inverting |
| Active site residues | known/not known |
| CAZy DB link | |
| http://www.cazy.org/GH174.html | |
Substrate specificities
Members of glycoside hydrolase family 174 have been shown to exhibit α-1,3-L-fucanase activity. The first member of this family, Fun174A from a marine bacterium Wenyingzhuangia aestuarii OF219, specifically hydrolyze the α-1,3- L-fucoside bonds between 2-O-sulfated and non-sulfated fucose residues in the sulfated fucan from sea cucumber Isostichopus badionotus in a processive endo-acting manner.[1] Meanwhile, three homologs of Fun174A from Rubritalea marina, Spartobacteria bacterium and Wenyingzhuangia fucanilytica, display activities toward sulfated fucan from Isostichopus badionotus.[1] All 92 members (up to May,2023) of GH174 are bacterial enzymes.
Kinetics and Mechanism
The catalytic mechanism of GH174 has not been identified. As mentioned in the report, Fun174A showed no transglycosylating activity in the tested acceptor substrates, such as glycerin, methanol, L-fucose, D-glucose, D-galactose, D-fructose, D-mannose, D-glucosamine and N-acetyl- D-glucosamine.[1]
Catalytic Residues
Multiple sequence alignments of GH174 homologs showed that D119, E120 and E218 in Fun174A were highly conserved in all sequences. Three single-site mutants D119E, E120A and E218Q were established, expressed and identified in the report. Mutant D119E, E120A and E218Q resulted in 100.0%, 85.7% and 88.3% loss of activity on sulfated fucan from Isostichopus badionotus respectively. It indicated that D119, E120 and E218 were critical for the functioning of Fun174A.[1]
Three-dimensional structures
No three-dimensional structure has been solved in this glycoside hydrolase family at present.
Family Firsts
- First stereochemistry determination
- Not yet identified.
- First catalytic nucleophile identification
- Not yet identified.
- First general acid/base residue identification
- Not yet identified.
- First 3-D structure
- Not yet identified.