CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Glycoside Hydrolase Family 19
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Vincent Eijsink^^^
- Responsible Curator: ^^^Vincent Eijsink^^^
| Glycoside Hydrolase Family GH19 | |
| Clan | none (lysozyme fold) |
| Mechanism | inverting |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/fam/GH19.html | |
Substrate specificities
Glycoside hydrolases of family 19 hydrolyze glycoside bonds in chitin, an insoluble polymer of beta-1,4-linked N-acetyl-D-glucosamine (GlcNAc) and are thus referred to as chitinases (EC 3.2.1.14). These enzymes were originally identified in plants. In an older classification system for plant chitinases, comprising both GH18 and GH19 chitinases, family 19 enzymes comprise classes I, II and IV. In 1996, the first bacterial family 19 chitinase was described [1]. In addition to cleaving chitin, GH19 chitinases cleave soluble oligomers of beta-1,4-linked N-acetyl-D-glucosamine. For some plant enzymes lysozyme activity has been demonstrated. Currently available data suggest that GH19 enzymes are not particularly effective in degrading crystalline chitin (compared to certain members of the GH18 chitinase family), especially enzymes that lack CBMs. On the other hand GH19 enzymes are highly active on chitosans (= partially deacetylated chitin) with high degrees of acetylation, even if they lack a CBM [2, 3]. Detailed studies on GH19 chitinases from Streptomyces (class IV; [3]) and rice (Oryza sativa; Class I; [4]) have revealed that productive binding requires a GlcNAc to be bound in subsites -2 and +1, whereas deacetylated GlcNAc (GlcN) is tolerated in subsites -1 and +2.
Kinetics and Mechanism
Family 19 enzymes employ an inverting mechanism, as determined by NMR [5] and HPLC [6]. Both structural characteristics (see below) and available biochemical data [3, 4] suggest that GH19 chitinases are non-processive endo-acting enzymes. Kinetic data for the conversion of polymeric and oligomeric substrates have been described in several studies. In some studies, kinetic data have been used to derive subsite binding affinties (e.g. [7, 8]).
Catalytic Residues
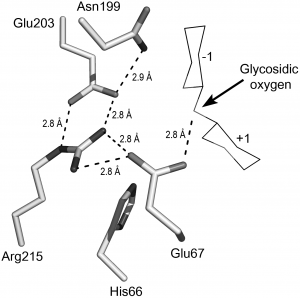
The catalytic residues are two glutamates. Although there still is limited structural information underpinning details of the inverting catalytic mechanism, there is considerable support for the notion that a glutamate located at the end of the third alpha helix (Glu 67 in the barley enzyme) acts as the catalytic acid, whereas another glutamate located in a more variable loop-like structure (Glu89 in the barley enzyme) acts as the catalytic base [9, 11, 12, 13].
It has been shown that at least two more conserved charged residues are crucial for catalysis. These residues, Glu203 and Arg215 in barley chitinase, form a triad together with the catalytic acid Glu67 [14] (see Figure). Interestingly, a similarly complex electrostatic interaction network is present in family GH46 chitosanases [15, 16] with whom the family 19 enzymes share some overall structural similarity (see below).
Three-dimensional structures
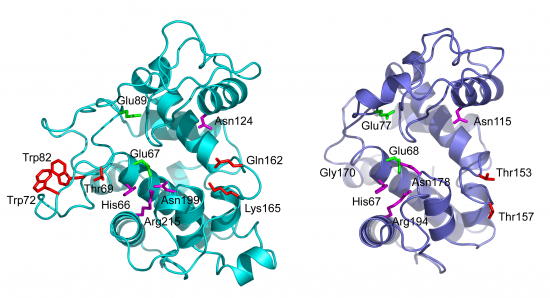
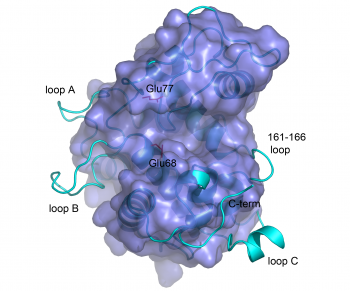
The catalytic domains of family 19 chitinases have a lysozyme-like fold with rather shallow substrate-binding grooves that are not particularly rich in aromatic residues (see Figure). The catalytic domains of family 19 chitinases share a common fold with family GH46 chitosanases and with lysozymes in families GH22, GH23 and GH24 of glycoside hydrolases [17]. For a long time, structural information for these chitinases was limited to the structures of two class II plant enzymes [11, 18]. Recently, the structures of bacterial family GH19 chitinases have become available ([13, 19]; class IV), as well as the structures of class I [20] and class IV [21] GH19 chitinases from plants.
The structures of bacterial GH19 chitinases revealed several differences from the previously reported plant structures ([13, 19]; see Figure). Compared to plant enzymes, the bacterial enzymes lack a C-terminal extension and three loops, some of which are thought to be flexible [20, 22].
There is no structural information for GH19 enzymes in complex with their substrate. In 2008, Huet et al published [9] the structure of a complex of papaya family 19 chitinase with GlcNAc units bound in the -2 and +1 subsites. This structure has been used to build a plausible model of a complex with (GlcNAc)4. This is the first structure (half experimental, half modeled) of an enzyme-substrate complex.
Family Firsts
- First primary sequence determination
- Bean leaf chitinase [23]
- First general base residue identification
- Chitinase from barley; determination by site-directed mutagenesis [12], structural analysis [11] and modelling [24]. Additional support from structure determination and modelling of a papaya chitinase [9].
- First general acid residue identification
- Chitinase from barley; determination by site-directed mutagenesis [12], structural analysis [11] and modelling [24]. Additional support from structure determination and modelling of a papaya chitinase [9].
- First 3-D structure
- Barley chitinase [11].
References
- Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M, and Watanabe T. (1996). A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol. 1996;178(17):5065-70. DOI:10.1128/jb.178.17.5065-5070.1996 |
- Kawase T, Yokokawa S, Saito A, Fujii T, Nikaidou N, Miyashita K, and Watanabe T. (2006). Comparison of enzymatic and antifungal properties between family 18 and 19 chitinases from S. coelicolor A3(2). Biosci Biotechnol Biochem. 2006;70(4):988-98. DOI:10.1271/bbb.70.988 |
- Heggset EB, Hoell IA, Kristoffersen M, Eijsink VG, and Vårum KM. (2009). Degradation of chitosans with chitinase G from Streptomyces coelicolor A3(2): production of chito-oligosaccharides and insight into subsite specificities. Biomacromolecules. 2009;10(4):892-9. DOI:10.1021/bm801418p |
- Sasaki C, Vårum KM, Itoh Y, Tamoi M, and Fukamizo T. (2006). Rice chitinases: sugar recognition specificities of the individual subsites. Glycobiology. 2006;16(12):1242-50. DOI:10.1093/glycob/cwl043 |
-
Fukamizo T, Koga D, Goto S Comparative biochemistry of chitinases - anomeric form of the reaction products Bioscience, Biotechnology & Biochemistry 1995, 59:311-313.
- Iseli B, Armand S, Boller T, Neuhaus JM, and Henrissat B. (1996). Plant chitinases use two different hydrolytic mechanisms. FEBS Lett. 1996;382(1-2):186-8. DOI:10.1016/0014-5793(96)00174-3 |
- Honda Y and Fukamizo T. (1998). Substrate binding subsites of chitinase from barley seeds and lysozyme from goose egg white. Biochim Biophys Acta. 1998;1388(1):53-65. DOI:10.1016/s0167-4838(98)00153-8 |
- Sasaki C, Itoh Y, Takehara H, Kuhara S, and Fukamizo T. (2003). Family 19 chitinase from rice (Oryza sativa L.): substrate-binding subsites demonstrated by kinetic and molecular modeling studies. Plant Mol Biol. 2003;52(1):43-52. DOI:10.1023/a:1023972007681 |
- Huet J, Rucktooa P, Clantin B, Azarkan M, Looze Y, Villeret V, and Wintjens R. (2008). X-ray structure of papaya chitinase reveals the substrate binding mode of glycosyl hydrolase family 19 chitinases. Biochemistry. 2008;47(32):8283-91. DOI:10.1021/bi800655u |
-
Hoell IA, Vaaje-Kolstad G, Eijsink VGH Structure and function of enzymes acting on chitin and chitosan Biotechnology and Genetic Engineering Reviews, Vol. 27, 2010, in press.
- Hart PJ, Monzingo AF, Ready MP, Ernst SR, and Robertus JD. (1993). Crystal structure of an endochitinase from Hordeum vulgare L. seeds. J Mol Biol. 1993;229(1):189-93. DOI:10.1006/jmbi.1993.1017 |
- Andersen MD, Jensen A, Robertus JD, Leah R, and Skriver K. (1997). Heterologous expression and characterization of wild-type and mutant forms of a 26 kDa endochitinase from barley (Hordeum vulgare L.). Biochem J. 1997;322 ( Pt 3)(Pt 3):815-22. DOI:10.1042/bj3220815 |
- Hoell IA, Dalhus B, Heggset EB, Aspmo SI, and Eijsink VG. (2006). Crystal structure and enzymatic properties of a bacterial family 19 chitinase reveal differences from plant enzymes. FEBS J. 2006;273(21):4889-900. DOI:10.1111/j.1742-4658.2006.05487.x |
- Ohnishi T, Juffer AH, Tamoi M, Skriver K, and Fukamizo T. (2005). 26 kDa endochitinase from barley seeds: an interaction of the ionizable side chains essential for catalysis. J Biochem. 2005;138(5):553-62. DOI:10.1093/jb/mvi154 |
- Fukamizo T, Juffer AH, Vogel HJ, Honda Y, Tremblay H, Boucher I, Neugebauer WA, and Brzezinski R. (2000). Theoretical calculation of pKa reveals an important role of Arg205 in the activity and stability of Streptomyces sp. N174 chitosanase. J Biol Chem. 2000;275(33):25633-40. DOI:10.1074/jbc.M002574200 |
- Lacombe-Harvey ME, Fukamizo T, Gagnon J, Ghinet MG, Dennhart N, Letzel T, and Brzezinski R. (2009). Accessory active site residues of Streptomyces sp. N174 chitosanase: variations on a common theme in the lysozyme superfamily. FEBS J. 2009;276(3):857-69. DOI:10.1111/j.1742-4658.2008.06830.x |
- Monzingo AF, Marcotte EM, Hart PJ, and Robertus JD. (1996). Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol. 1996;3(2):133-40. DOI:10.1038/nsb0296-133 |
- Hahn M, Hennig M, Schlesier B, and Höhne W. (2000). Structure of jack bean chitinase. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 9):1096-9. DOI:10.1107/s090744490000857x |
- Kezuka Y, Ohishi M, Itoh Y, Watanabe J, Mitsutomi M, Watanabe T, and Nonaka T. (2006). Structural studies of a two-domain chitinase from Streptomyces griseus HUT6037. J Mol Biol. 2006;358(2):472-84. DOI:10.1016/j.jmb.2006.02.013 |
- Ubhayasekera W, Tang CM, Ho SWT, Berglund G, Bergfors T, Chye ML, and Mowbray SL. (2007). Crystal structures of a family 19 chitinase from Brassica juncea show flexibility of binding cleft loops. FEBS J. 2007;274(14):3695-3703. DOI:10.1111/j.1742-4658.2007.05906.x |
- Ubhayasekera W, Rawat R, Ho SW, Wiweger M, Von Arnold S, Chye ML, and Mowbray SL. (2009). The first crystal structures of a family 19 class IV chitinase: the enzyme from Norway spruce. Plant Mol Biol. 2009;71(3):277-89. DOI:10.1007/s11103-009-9523-9 |
- Fukamizo T, Miyake R, Tamura A, Ohnuma T, Skriver K, Pursiainen NV, and Juffer AH. (2009). A flexible loop controlling the enzymatic activity and specificity in a glycosyl hydrolase family 19 endochitinase from barley seeds (Hordeum vulgare L.). Biochim Biophys Acta. 2009;1794(8):1159-67. DOI:10.1016/j.bbapap.2009.03.009 |
- Broglie KE, Gaynor JJ, and Broglie RM. (1986). Ethylene-regulated gene expression: molecular cloning of the genes encoding an endochitinase from Phaseolus vulgaris. Proc Natl Acad Sci U S A. 1986;83(18):6820-4. DOI:10.1073/pnas.83.18.6820 |
- Brameld KA and Goddard WA 3rd. (1998). The role of enzyme distortion in the single displacement mechanism of family 19 chitinases. Proc Natl Acad Sci U S A. 1998;95(8):4276-81. DOI:10.1073/pnas.95.8.4276 |