CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 52"
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
{{UnderConstruction}} | {{UnderConstruction}} | ||
| − | * [[Author]]: ^^^Julie Grondin^^^ | + | * [[Author]]: ^^^Julie Grondin^^^, ^^^Brian Lowrance^^^ |
* [[Responsible Curator]]: | * [[Responsible Curator]]: | ||
---- | ---- | ||
| Line 29: | Line 29: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | GH52 enzymes are | + | The GH52 enzymes are often isolated from various mesophilic and thermophilic bacteria, which has led to a demonstrated high thermostability within this family. The enzymes are monospecific, functioning as ''exo''-β-xylosidases (EC [{{EClink}}3.2.1.37 3.2.1.37]) that cleave the terminal xylose residues from the nonreducing end of xylooligosaccharides, such as ''p''NP-β-d-xylopyranoside <cite>Bravman2001, Espina2014</cite>, xylobiose <cite>Espina2014</cite>, xylotriose <cite>Espina2014</cite>. Low levels of α-L-arabinofuranoside activity has also been observed within members of the GH52 family <cite>Suzuki2014, Bravman2003a</cite>, which is similar to the specificity noted for GH13 and GH54 families for β-xylooligosaccharides and α-L-arabinofuranoside. The specificity for these substrates is likely due to similarities in orientation of hydroxyls and glycosidic bonds of the substrate within the active site <cite>Lee2007, Utt1991</cite>. The plasticity of the active site of some GH52 members has been further explored through site-directed mutagenesis, where introduction of xylanase activity <cite>Huang2014</cite> and transition from a glycosyl hydrolase to a glycosynthase <cite>Dann2014</cite> has been achieved. Some enzymes in the family have also exhibited weak transglycosylation activity, a phenomenon that has been infrequently observed in other glycosyl hydrolases as well <cite>Romero2019</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | GH52 | + | Retention of stereochemistry has been observed in GH52 β-xylosidases, characteristic of a classical [[Koshland double-displacement mechanism]] <cite>Koshland1953</cite>. This was first determined by Bravmen, et. al using H-NMR to analyze the breakdown products of ''p''NP-β-D-xylopyranoside by XynB2, a β-xylosidase, from ''Bacillus stearothermophilus'' T-6 <cite>Bravman2001</cite>. The first detailed kinetic analysis within this family was published in 2003 by Bravman, et. al, containing standard kinetics (''p''NP xylobiose k<sub>cat</sub>/K<sub>m</sub>= 140 s<sup>-1</sup>mM<sup>-1</sup>), pH dependence (enzymatic catalysis is dependent the ionizable residues E335 and D495, with free enzyme experimental pKa values of 4.2 and 7.3, respectively) and substrate binding kinetics (xylobiose 17.1x10<sup>4</sup> M<sup>-1</sup>, xylotriose 9.6x10<sup>4</sup> M-<sup>1</sup>) of the ''exo''-β-xylosidase XynB2 from ''Bacillus stearothermophilus'', among other kinetic parameters <cite>Bravman2003a</cite>. |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | Site-directed mutagenesis, chemical rescue, and kinetic profiling of XynB2 from | + | Site-directed mutagenesis, chemical rescue, and kinetic profiling of XynB2 from ''B. stearothermophilus'' T-6 identified E335 as the [[catalytic nucleophile]], and D495 as the [[general acid/base]] <cite>Bravman2001, Bravman2003b</cite>. The catalytic nucleophile (E335) is conserved within the WVVNEGEY motif, which is found approximately 150 residues up-stream from the EITTYDSLD motif containing the general acid/base (D495). These results were further confirmed following the structural analysis of GH52 from ''Geobacillus thermoglucosidasius'' <cite>Espina2014</cite>, the 6.5Å separation of Glu and Asp in the active site of this enzyme is consistent with other retaining enzymes. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | [[File:Figure1_dimer.PNG| | + | [[File:Figure1_dimer.PNG|300px|thumb|right|'''Figure 1. The dimeric structure of GH52 from ''Geobacillus thermoglucosidasius'' in complex with xylobiose (orange)([{{PDBlink}}4C1P PDB ID 4C1P]).''' The active site is enclosed by residues from both monomers, restricting this enzyme to ''exo''-hydrolysis via steric hindrance of the catalytic site. Figure from <cite>Espina2014</cite>.]] |
| − | |||
| − | The '' | + | The structure of GH52 consists of an N-terminal β-sandwich domain and a C-terminal (α/α)<sup>6</sup> barrel domain, classifying these enzymes into the GH-''O'' clan, similar to that noted for the GH116 family. The exo-acting mode of action of GH52 is reflected in the topology of the active site. The enzyme acts as a dimer in solution <cite>Bravman2001, Espina2014</cite>, with interactions between monomers forming a deep pocket to enclose and distort the non-reducing end xylose into a high-energy <sup>4</sup>H<sub>3</sub> half-chair transition conformation, while simultaneously hindering the entry of large xylan polymers into the catalytic site <cite>Espina2014</cite>. Furthermore, the structure of the active site allosterically inhibits access to negative subsites beyond the -1 site. This permits interaction with only a single xylosyl residue in the negative subsites and thus hydrolysis yields a lone xylose molecule. This mechanism promotes strict ''exo''-β-xylosidase activity while inhibiting activity on large polymers such as xylan. |
== Family Firsts == | == Family Firsts == | ||
| Line 50: | Line 49: | ||
== References == | == References == | ||
| + | |||
<biblio> | <biblio> | ||
#Bravman2001 pmid=11322943 | #Bravman2001 pmid=11322943 | ||
#Espina2014 pmid=24816105 | #Espina2014 pmid=24816105 | ||
| + | #Suzuki2001 pmid=11330658 | ||
| + | #Bravman2003a pmid=12950180 | ||
| + | #Lee2007 pmid=18051350 | ||
| + | #Utt1991 pmid=1905520 | ||
| + | #Huang2014 pmid=24122394 | ||
| + | #Dann2014 pmid=25484225 | ||
| + | #Romero2019 pmid=31024890 | ||
| + | #Bravman2003b pmid=12738774 | ||
#Koshland1953 Koshland DE Jr: Stereochemistry and the mechanism of enzyme reactions. Biol Rev 1953, 28:416-436. [https://doi.org/10.1111/j.1469-185X.1953.tb01386.x DOI:10.1111/j.1469-185X.1953.tb01386.x] | #Koshland1953 Koshland DE Jr: Stereochemistry and the mechanism of enzyme reactions. Biol Rev 1953, 28:416-436. [https://doi.org/10.1111/j.1469-185X.1953.tb01386.x DOI:10.1111/j.1469-185X.1953.tb01386.x] | ||
| − | + | ||
</biblio> | </biblio> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
[[Category:Glycoside Hydrolase Families|GH052]] | [[Category:Glycoside Hydrolase Families|GH052]] | ||
Revision as of 14:38, 3 September 2020
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Julie Grondin^^^, ^^^Brian Lowrance^^^
- Responsible Curator:
| Glycoside Hydrolase Family GH52 | |
| Clan | GH-O |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH52.html | |
Substrate specificities
The GH52 enzymes are often isolated from various mesophilic and thermophilic bacteria, which has led to a demonstrated high thermostability within this family. The enzymes are monospecific, functioning as exo-β-xylosidases (EC 3.2.1.37) that cleave the terminal xylose residues from the nonreducing end of xylooligosaccharides, such as pNP-β-d-xylopyranoside [1, 2], xylobiose [2], xylotriose [2]. Low levels of α-L-arabinofuranoside activity has also been observed within members of the GH52 family [3, 4], which is similar to the specificity noted for GH13 and GH54 families for β-xylooligosaccharides and α-L-arabinofuranoside. The specificity for these substrates is likely due to similarities in orientation of hydroxyls and glycosidic bonds of the substrate within the active site [5, 6]. The plasticity of the active site of some GH52 members has been further explored through site-directed mutagenesis, where introduction of xylanase activity [7] and transition from a glycosyl hydrolase to a glycosynthase [8] has been achieved. Some enzymes in the family have also exhibited weak transglycosylation activity, a phenomenon that has been infrequently observed in other glycosyl hydrolases as well [9].
Kinetics and Mechanism
Retention of stereochemistry has been observed in GH52 β-xylosidases, characteristic of a classical Koshland double-displacement mechanism [10]. This was first determined by Bravmen, et. al using H-NMR to analyze the breakdown products of pNP-β-D-xylopyranoside by XynB2, a β-xylosidase, from Bacillus stearothermophilus T-6 [1]. The first detailed kinetic analysis within this family was published in 2003 by Bravman, et. al, containing standard kinetics (pNP xylobiose kcat/Km= 140 s-1mM-1), pH dependence (enzymatic catalysis is dependent the ionizable residues E335 and D495, with free enzyme experimental pKa values of 4.2 and 7.3, respectively) and substrate binding kinetics (xylobiose 17.1x104 M-1, xylotriose 9.6x104 M-1) of the exo-β-xylosidase XynB2 from Bacillus stearothermophilus, among other kinetic parameters [4].
Catalytic Residues
Site-directed mutagenesis, chemical rescue, and kinetic profiling of XynB2 from B. stearothermophilus T-6 identified E335 as the catalytic nucleophile, and D495 as the general acid/base [1, 11]. The catalytic nucleophile (E335) is conserved within the WVVNEGEY motif, which is found approximately 150 residues up-stream from the EITTYDSLD motif containing the general acid/base (D495). These results were further confirmed following the structural analysis of GH52 from Geobacillus thermoglucosidasius [2], the 6.5Å separation of Glu and Asp in the active site of this enzyme is consistent with other retaining enzymes.
Three-dimensional structures
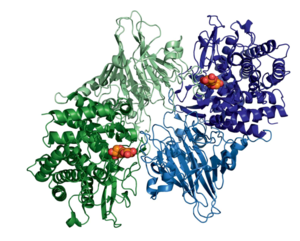
The structure of GH52 consists of an N-terminal β-sandwich domain and a C-terminal (α/α)6 barrel domain, classifying these enzymes into the GH-O clan, similar to that noted for the GH116 family. The exo-acting mode of action of GH52 is reflected in the topology of the active site. The enzyme acts as a dimer in solution [1, 2], with interactions between monomers forming a deep pocket to enclose and distort the non-reducing end xylose into a high-energy 4H3 half-chair transition conformation, while simultaneously hindering the entry of large xylan polymers into the catalytic site [2]. Furthermore, the structure of the active site allosterically inhibits access to negative subsites beyond the -1 site. This permits interaction with only a single xylosyl residue in the negative subsites and thus hydrolysis yields a lone xylose molecule. This mechanism promotes strict exo-β-xylosidase activity while inhibiting activity on large polymers such as xylan.
Family Firsts
- First stereochemistry determination
- XynB2 from Bacillus stearothermophilus T-6 by 1H-NMR for the hydrolysis of pNP-β-D-xylopyranoside [1].
- First catalytic nucleophile identification
- XynB2 from Bacillus stearothermophilus T-6 by site-directed mutagenesis and chemical rescue [12].
- First general acid/base residue identification
- XynB2 from Bacillus stearothermophilus T-6 by site-directed mutagenesis, chemical rescue, and pH profiling [12].
- First 3-D structure
- GH52 from Geobacillus thermoglucosidasius NBRC 107763 [2].
References
- Bravman T, Zolotnitsky G, Shulami S, Belakhov V, Solomon D, Baasov T, Shoham G, and Shoham Y. (2001). Stereochemistry of family 52 glycosyl hydrolases: a beta-xylosidase from Bacillus stearothermophilus T-6 is a retaining enzyme. FEBS Lett. 2001;495(1-2):39-43. DOI:10.1016/s0014-5793(01)02360-2 |
- Espina G, Eley K, Pompidor G, Schneider TR, Crennell SJ, and Danson MJ. (2014). A novel β-xylosidase structure from Geobacillus thermoglucosidasius: the first crystal structure of a glycoside hydrolase family GH52 enzyme reveals unpredicted similarity to other glycoside hydrolase folds. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 5):1366-74. DOI:10.1107/S1399004714002788 |
- Bravman T, Zolotnitsky G, Belakhov V, Shoham G, Henrissat B, Baasov T, and Shoham Y. (2003). Detailed kinetic analysis of a family 52 glycoside hydrolase: a beta-xylosidase from Geobacillus stearothermophilus. Biochemistry. 2003;42(35):10528-36. DOI:10.1021/bi034505o |
- Lee TH, Lim PO, and Lee YE. (2007). Cloning, characterization, and expression of xylanase A gene from Paenibacillus sp. DG-22 in Escherichia coli. J Microbiol Biotechnol. 2007;17(1):29-36. | Google Books | Open Library
- Utt EA, Eddy CK, Keshav KF, and Ingram LO. (1991). Sequencing and expression of the Butyrivibrio fibrisolvens xylB gene encoding a novel bifunctional protein with beta-D-xylosidase and alpha-L-arabinofuranosidase activities. Appl Environ Microbiol. 1991;57(4):1227-34. DOI:10.1128/aem.57.4.1227-1234.1991 |
- Huang Z, Liu X, Zhang S, and Liu Z. (2014). GH52 xylosidase from Geobacillus stearothermophilus: characterization and introduction of xylanase activity by site‑directed mutagenesis of Tyr509. J Ind Microbiol Biotechnol. 2014;41(1):65-74. DOI:10.1007/s10295-013-1351-x |
- Dann R, Lansky S, Lavid N, Zehavi A, Belakhov V, Baasov T, Dvir H, Manjasetty B, Belrhali H, Shoham Y, and Shoham G. (2014). Preliminary crystallographic analysis of Xyn52B2, a GH52 β-D-xylosidase from Geobacillus stearothermophilus T6. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 12):1675-82. DOI:10.1107/S2053230X14023887 |
- Romero-Téllez S, Lluch JM, González-Lafont À, and Masgrau L. (2019). Comparing Hydrolysis and Transglycosylation Reactions Catalyzed by Thermus thermophilus β-Glycosidase. A Combined MD and QM/MM Study. Front Chem. 2019;7:200. DOI:10.3389/fchem.2019.00200 |
-
Koshland DE Jr: Stereochemistry and the mechanism of enzyme reactions. Biol Rev 1953, 28:416-436. DOI:10.1111/j.1469-185X.1953.tb01386.x
- Bravman T, Belakhov V, Solomon D, Shoham G, Henrissat B, Baasov T, and Shoham Y. (2003). Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus stearothermophilus T-6. J Biol Chem. 2003;278(29):26742-9. DOI:10.1074/jbc.M304144200 |
- Suzuki T, Kitagawa E, Sakakibara F, Ibata K, Usui K, and Kawai K. (2001). Cloning, expression, and characterization of a family 52 beta-xylosidase gene (xysB) of a multiple-xylanase-producing bacterium, Aeromonas caviae ME-1. Biosci Biotechnol Biochem. 2001;65(3):487-94. DOI:10.1271/bbb.65.487 |