CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 99"
| Line 28: | Line 28: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | [[Image:GH99_substrates.png|thumb|right|300px|'''Figure 1. Endo-α-mannosidase is located in the Golgi apparatus and pre-Golgi intermediates and acts on folded and unfolded ER-escaped glucosylated N-linked glycoproteins (X = protein); glucosylated free oligosaccharides (X = OH); and dolichol-bound glucosylated glycans (X = diphosphodolichol).''']] | + | [[Image:GH99_substrates.png|thumb|right|300px|'''Figure 1. (Top) Endo-α-mannosidase is located in the Golgi apparatus and pre-Golgi intermediates and acts on folded and unfolded ER-escaped glucosylated N-linked glycoproteins (X = protein); glucosylated free oligosaccharides (X = OH); and dolichol-bound glucosylated glycans (X = diphosphodolichol). (Bottom) Endo-α-1,2-mannanases are bacterial enzymes with the ability to cleave Man<sub>3</sub>Man<sub>9</sub>GlcNAc<sub>2</sub> substructures within fungal cell wall mannan at the indicated position, releasing α-1,3-mannobiose.''']] |
Revision as of 19:30, 8 December 2014
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Spencer Williams^^^
- Responsible Curator: ^^^Gideon Davies^^^
| Glycoside Hydrolase Family GH99 | |
| Clan | not assigned |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH99.html | |
Substrate specificities
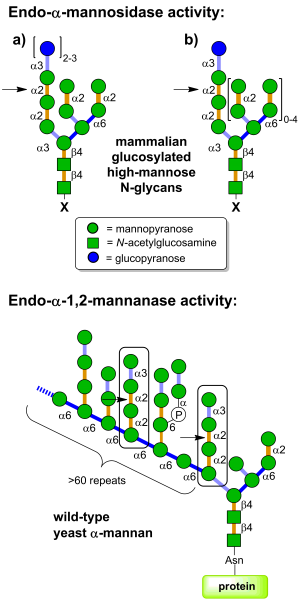
Glycoside hydrolases of family GH99 are endo-acting enzymes. This family was originally created from the mammalian enzyme, cloned by Spiro and co-workers [1]. Two main activities have been described for this family. Endo-α-mannosidase activity describes the capacity to cleave glucose-substituted mannose within immature N-linked glycans of the general formula Glc1-3Man9GlcNAc2 (or structures trimmed in the B and C branches). On the other hand endo-α-1,2-mannanase activity describes the ability to cleave αMan-
1,3-αMan-1,2-αMan-1,2-αMan sequences within fungal cell wall mannans, releasing α-1,3-mannobiose [2].
Mammalian endo-α-mannosidases typically possess maximal activity on the monoglucosylated forms [3]. Mammalian endo-α-mannosidases are localized to the Golgi apparatus [4] and appear to play a role in the rescue of glucosylated N-linked glycans that have evaded the action of the endoplasmic reticulum exo-glucosidases I and II [5]. Mammalian endo-α-mannosidase has greater activity on glucosylated N-linked glycans that have been trimmed in the non-glucose-substituted branches [1]. There is evidence that mammalian endo-α-mannosidases act on dolichol-bound N-glycan precursors [6], as well as free oligosaccharides released from N-glycoproteins and which undergo retrograde transport through the secretory pathway [7]. A fluorescent tetrasaccharide fragment (Glcα1-3Manα1-2Manα1-2Manα1-O-C3H6-NH-Dansyl) has been developed that is a substrate for mammalian endo-α-mannosidase [8]. Studies of bacterial GH99 enzymes from Shewanella amazonensis [9], Bacteroides thetaiotaomicron and Bacteroides xylanisolvens [10] have shown that these enzymes can also process Glc1/3Man9/7GlcNAc2 structures, matching the substrate specificity of the mammalian enzymes. Kinetic analysis of Bacteroides thetaiotaomicron GH99 on a fluorescently-labelled Glc3Man9GlcNAc2 structure yielded kinetic parameters that were similar to that found for the mammalian enzyme [10]. Both mammalian and bacterial enzymes can utilize simple 'reducing end' substrate mimics such as methylumbelliferyl α-glucosyl-1,3-α-mannopyranoside [11] or α-glucosyl-1,3-α-mannopyranosyl fluoride [10], but are inactive on aryl mannosides.
Bacterial endo-α-1,2-mannanases from B. thetaiotaomicron and B. xylanisolvens have superior kinetics for cleavage of Man3Man9GlcNAc2-OMe versus glucosylated high mannose N-glycans [2]. These two enzymes displayed a 10-fold preference for methylumbelliferyl α-mannosyl-1,3-α-mannopyranoside versus methylumbelliferyl α-glucosyl-1,3-α-mannopyranoside [2].
Kinetics and Mechanism
Family GH99 endo-α-mannosidases are retaining enzymes, as first shown by 1H NMR analysis of the hydrolysis of α-glucosyl-1,3-α-mannopyranosyl fluoride by Bacteroides thetaiotaomicron [10]. Retaining mannoside hydrolases (eg GH38) typically follow a classical Koshland double-displacement mechanism. However, X-ray crystallographic analysis of BxGH99 and BtGH99 failed to reveal a candidate nucleophilic residue; instead an alternative mechanism involving substrate-assisted catalysis by the 2OH residue and proceeding through a 1,2-anhydro sugar was proposed [10]. In this proposal, Glu333 in BxGH99 (Glu329 in BtGH99) acts as a general acid/base to deprotonate the 2OH and protonate the 1,2-anhydrosugar.
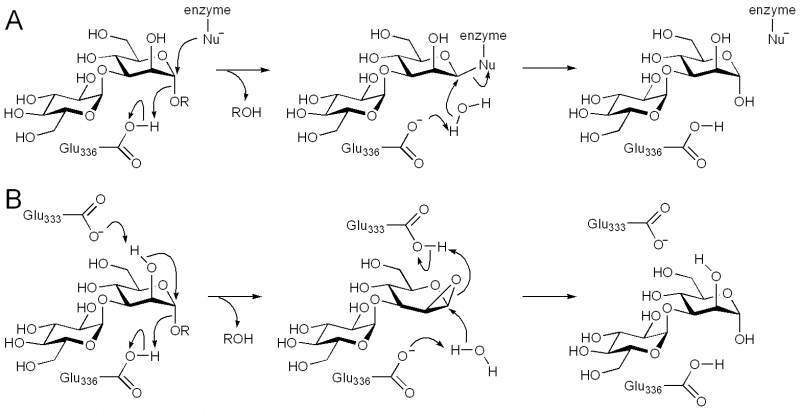
Catalytic Residues
The general acid/base was highlighted by X-ray crystallographic analysis as Glu336 in BxGH99 and Glu332 in BtGH99 [10]. The Glu332Ala mutant of BtGH99 shows a 50-fold decrease in catalytic activity relative to the wild-type enzyme using the activated substrate α-glucosyl-1,3-α-mannopyranosyl fluoride, and undetectable activity against the natural substrate, Glc3Man7GlcNAc2, supporting the role of this residue as general acid/base. As described in "Kinetics and Mechanism" the identity of the nucleophilic residue used for catalysis is more obscure and the 2OH of the substrate has been proposed to act as a nucleophile in a mechanism involving substrate assisted catalysis [10]. According to this proposal, Glu333 in BxGH99 (Glu329 in BtGH99) acts as a general acid/base to deprotonate the 2OH and protonate the 1,2-anhydrosugar.
Three-dimensional structures
Three-dimensional structures are available for bacterial members of GH99, including B. thetaiotaomicron and B. xylanisolvens [10]. They have a classical (β/α)8 TIM barrel fold, which is distinguished by the presence of extended loop motifs that form the active site. In different structures of the bacterial enzymes, these loops adopt different conformations, and appear to play a role in recognizing the extended structure of the N-glycan substrate. Binary complexes with two inhibitors, α-glucosyl-1,3-deoxymannonojirimycin and α-glucosyl-1,3-isofagomine, and 'active-site-spanning' ternary complexes with the same two inhibitors and the reducing end product fragment 1,2-α-mannobiose, provided insight into active site residues, especially the acid/base (Glu336 in BxGH99; Glu332 in BtGH99) and another key residue that interacts with both the 2OH of the -1 mannose residue and the 6OH of the -2 glucose residue, which provides a rationale for the requirement of a glucosylated-mannoside as the minimal substrate for GH99 enzymes [10]. As discussed in more detail in the "Kinetics and Mechanism" section, the precise identity of the nucleophilic residue is unclear, as in all GH99-inhibitor complexes with an occupied -1 subsite there is no candidate nucleophile within a reasonable distance to the "anomeric" carbon: in BxGH99 Glu333 is approximately 3.5 Å distant and the OH of Tyr46 and Tyr252 are 4.0 Å distant.
Comparison of binary complexes of α-mannosyl-1,3-isofagomine and α-glucosyl-1,3-isofagomine with B. xylanisolvens GH99 revealed almost identical binding [2]. It was speculated that a key residue in the -2 subsite, tryptophan-126 in BxGH99, provided a preference for mannose-configured substrates in these bacterial endo-α-1,2-mannanases. By contrast the equivalent residue in mammalian endo-α-mannosidases is a highly conserved tyrosine, which it was speculated may result in a favorable hydrogen bond to a glucose-configured substrate and confer endo-α-mannosidase activity.
Sample structures
| Three-dimensional structure of GH99 endo-α-mannosidase from Bacteroides xylanisolvens, PDB code [1] [10]. | Three-dimensional structure of GH99 endo-α-mannosidase from Bacteroides xylanisolvens bound to glucose-1,3-isofagomine and α-1,2- mannobiose, PDB code [2] [10]. |
|---|---|
|
<jmol> <jmolApplet> <color>white</color> <frame>true</frame> <uploadedFileContents>4ad1.pdb</uploadedFileContents> <script>cpk off; wireframe off; cartoon; color cartoon powderblue; select ligand; wireframe 0.3; select MG; spacefill; set spin Y 10; spin off; set antialiasDisplay OFF</script> </jmolApplet> </jmol> |
<jmol> <jmolApplet> <color>white</color> <frame>true</frame> <uploadedFileContents>4ad4.pdb</uploadedFileContents> <script>cpk off; wireframe off; cartoon; color cartoon powderblue; select GLC or IFG; wireframe 0.3; select MAN; wireframe 0.3; set spin Y 10; spin off; set antialiasDisplay OFF</script> </jmolApplet> </jmol> |
Family Firsts
- First sterochemistry determination
- Bacteroides thetaiotaomicron endo-α-mannosidase by 1H NMR [10]
- First catalytic nucleophile identification
- It has been proposed that GH99 enzymes operate through a mechanism involving substrate assisted catalysis by the 2OH group of the -1 mannose residue [10]
- First general acid/base residue identification
- Bacteroides thetaiotaomicron endo-α-mannosidase by X-ray crystallography and analysis of enzyme mutant activities [10]
- First 3-D structure of a GH99 enzyme
- Bacteroides thetaiotaomicron and Bacteroides xylanisolvens endo-α-mannosidases [10]
References
- Spiro MJ, Bhoyroo VD, and Spiro RG. (1997). Molecular cloning and expression of rat liver endo-alpha-mannosidase, an N-linked oligosaccharide processing enzyme. J Biol Chem. 1997;272(46):29356-63. DOI:10.1074/jbc.272.46.29356 |
- Roth J, Ziak M, and Zuber C. (2003). The role of glucosidase II and endomannosidase in glucose trimming of asparagine-linked oligosaccharides. Biochimie. 2003;85(3-4):287-94. DOI:10.1016/s0300-9084(03)00049-x |
- Zuber C, Spiro MJ, Guhl B, Spiro RG, and Roth J. (2000). Golgi apparatus immunolocalization of endomannosidase suggests post-endoplasmic reticulum glucose trimming: implications for quality control. Mol Biol Cell. 2000;11(12):4227-40. DOI:10.1091/mbc.11.12.4227 |
- Dale MP, Kopfler WP, Chait I, and Byers LD. (1986). Beta-glucosidase: substrate, solvent, and viscosity variation as probes of the rate-limiting steps. Biochemistry. 1986;25(9):2522-9. DOI:10.1021/bi00357a036 |
- Spiro MJ and Spiro RG. (2000). Use of recombinant endomannosidase for evaluation of the processing of N-linked oligosaccharides of glycoproteins and their oligosaccharide-lipid precursors. Glycobiology. 2000;10(5):521-9. DOI:10.1093/glycob/10.5.521 |
- Kukushkin NV, Alonzi DS, Dwek RA, and Butters TD. (2011). Demonstration that endoplasmic reticulum-associated degradation of glycoproteins can occur downstream of processing by endomannosidase. Biochem J. 2011;438(1):133-42. DOI:10.1042/BJ20110186 |
- Iwamoto S, Kasahara Y, Kamei K, Seko A, Takeda Y, Ito Y, and Matsuo I. (2014). Measurement of endo-α-mannosidase activity using a fluorescently labeled oligosaccharide derivative. Biosci Biotechnol Biochem. 2014;78(6):927-36. DOI:10.1080/09168451.2014.910101 |
- Matsuda K, Kurakata Y, Miyazaki T, Matsuo I, Ito Y, Nishikawa A, and Tonozuka T. (2011). Heterologous expression, purification, and characterization of an α-mannosidase belonging to glycoside hydrolase family 99 of Shewanella amazonensis. Biosci Biotechnol Biochem. 2011;75(4):797-9. DOI:10.1271/bbb.100874 |
- Thompson AJ, Williams RJ, Hakki Z, Alonzi DS, Wennekes T, Gloster TM, Songsrirote K, Thomas-Oates JE, Wrodnigg TM, Spreitz J, Stütz AE, Butters TD, Williams SJ, and Davies GJ. (2012). Structural and mechanistic insight into N-glycan processing by endo-α-mannosidase. Proc Natl Acad Sci U S A. 2012;109(3):781-6. DOI:10.1073/pnas.1111482109 |
-
Vogel C, Pohlentz G. Synthesis of α-D-glucopyranosyl-(1,3)-α-D-mannopyranosyl-(1,7)-4-methylumbelliferone, a fluorogenic substrate for endo-α-1,2-mannosidase. J. Carbohydr. Chem. 2000, 19: 1247-1258.