CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 146
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH146 | |
| Clan | GH-A |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH146.html | |
Substrate specificities
Enzymes of this family of glycoside hydrolases exhibit β-arabinofuranosidase activity. The founding member of this family, BT0349 from Bacteroides thetaiotaomicron, cleaves both β1,2- and β1,3-linked arabinofuranose side chains present in branched sugar beet arabinan [1].
Kinetics and Mechanism
BT0349 β-arabinofuranosidase displays exo-activity on β-linked arabinofuranosyl groups and has been proposed to act with a retaining mechanism, based on the positions of the catalytic residues (see below) [1].
Catalytic Residues
The catalytic nucleophile and general acid/base residues of BT0349 are proposed to comprise Cys414 and Glu318, respectively [1]. This is based on the conservation of these residues with the catalytic apparatus of GH127 β-arabinofuranosidases [2].
Three-dimensional structures
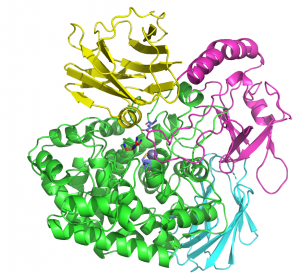
The crystal structure of BT0349, solved using single-wavelength anomalous dispersion methods to a resolution of 2.1 A, revealed a four-domain structure. The N-terminal catalytic domain comprises an (α/α)6 barrel followed by three β-sandwich domains (1, 2 & 3). Arabinofuranose was present in the active site pocket of the catalytic domain, while a zinc atom is coordinated by three cysteine residues and a glutamate in the same domain [1]. The catalytic apparatus is conserved with a GH127 β-arabinofuranosidase [2]; however, the GH127 enzyme lacks β-sandwich domain 3, which is positioned over the active site, effectively burying the bound arabinofuranose. GH127 enzymes were shown to cleave glycosidic bonds with retention of anomeric configuration [3] and, given the conservation of the catalytic apparatus in GH127 and GH146 members, enzymes in both families were proposed to hydrolyse arabinofururanosidic bonds through a retaining mechanism [1].
Family Firsts
- First stereochemistry determination
- Retaining mechanism of the B. thetaiotaomicron β-arabinofuranosidase, BT0349 [1], based on conservation of the catalytic apparatus with retaining GH127 enzymes [2, 3].
- First catalytic nucleophile identification
- Cys414 of the B. thetaiotaomicron β-arabinofuranosidase, BT0349, based on mutagenesis data, 3D structure [1] and conservation with the catalytic nucleophile of GH127 enzymes [2].
- First general acid/base residue identification
- Glu318 of the B. thetaiotaomicron β-arabinofuranosidase, BT0349, based on mutagenesis data, 3D structure [1]and conservation with the catalytic nucleophile of GH127 enzymes [2].
- First 3-D structure
- The B. thetaiotaomicron β-arabinofuranosidase, BT0349. The enzyme contains an N-terminal catalytic domain that folds into a (α/α)6 barrel, which is followed by three β-sandwich domains.
References
- Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet MC, Henrissat B, Martens EC, Mosimann SC, Abbott DW, and Gilbert HJ. (2018). Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol. 2018;3(2):210-219. DOI:10.1038/s41564-017-0079-1 |
- Ito T, Saikawa K, Kim S, Fujita K, Ishiwata A, Kaeothip S, Arakawa T, Wakagi T, Beckham GT, Ito Y, and Fushinobu S. (2014). Crystal structure of glycoside hydrolase family 127 β-l-arabinofuranosidase from Bifidobacterium longum. Biochem Biophys Res Commun. 2014;447(1):32-7. DOI:10.1016/j.bbrc.2014.03.096 |
- Fujita K, Takashi Y, Obuchi E, Kitahara K, and Suganuma T. (2014). Characterization of a novel β-L-arabinofuranosidase in Bifidobacterium longum: functional elucidation of a DUF1680 protein family member. J Biol Chem. 2014;289(8):5240-9. DOI:10.1074/jbc.M113.528711 |