CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Esterase Family 2"
Joel Weadge (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (73 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
{{CuratorApproved}} | {{CuratorApproved}} | ||
| − | * [[Author]]: | + | * [[Author]]: [[User:Bobby Lamont|Bobby Lamont]] |
| − | * [[Responsible Curator]]s: | + | * [[Responsible Curator]]s: [[User:Anthony Clarke|Anthony Clarke]] and [[User:Joel Weadge|Joel Weadge]] |
---- | ---- | ||
| Line 11: | Line 11: | ||
|{{Hl2}} colspan="2" align="center" |'''Carbohydrate Esterase Family CE2''' | |{{Hl2}} colspan="2" align="center" |'''Carbohydrate Esterase Family CE2''' | ||
|- | |- | ||
| − | |''' | + | |'''Fold''' |
|α/β-hydrolase | |α/β-hydrolase | ||
|- | |- | ||
| Line 18: | Line 18: | ||
|- | |- | ||
|'''Active site residues''' | |'''Active site residues''' | ||
| − | |Known | + | |Known, Catalytic Dyad |
|- | |- | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
| Line 28: | Line 28: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | + | All of the well characterized carbohydrate esterase family 2 enzymes have been shown to remove acetate groups from the synthetic molecule, 4-nitrophenyl acetate (''p''NP-Ac) <cite>Montanier2009 Till2013</cite>. CE2 family members have also demonstrated preferential de-''O''-acetylation of xylopyranosides at positions 3 and 4, over the 2 position. In expanded substrate profiles, CE2 enzymes were also noted to deacetylate glucopyranosyl and mannopyranosyl residues at the 6-''O'' position. The greater catalytic activity when deacetylating mannopyranosyl and glucopyranosyl compared to xylopyranosides has prompted the naming of some CE2 family members as 6-de-''O''-acetylases <cite>Topakas2010</cite>. | |
| − | All of the well characterized carbohydrate esterase family 2 enzymes have been shown to remove acetate groups from the synthetic molecule, 4-nitrophenyl acetate (''p''NP-Ac) <cite>Montanier2009 Till2013</cite>. | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | + | Most CE2 family members contain a catalytic dyad <cite>Montanier2009</cite>. For example, the structurally characterized ''Ct''CE2 ([{{PDBlink}}2WAO PDB ID 2WAO]) from ''Clostridium thermocellum'', ''Cj''CE2A ([{{PDBlink}}2WAA PDB ID 2WAA]) from ''Cellvibrio japonicus'', and Est2A ([{{PDBlink}}3U37 PDB ID 3U37]) from ''Butyrivibrio proteoclasticus'' contain conserved serine and histidine residues that form the catalytic dyad and lack a third aspartate residue that is typically found in serine esterase triads <cite>Montanier2009 Till2013</cite>. Without the aspartate residue, the histidine of the catalytic dyads are supported by main-chain carbonyl groups provided by a backbone amino acid. In cases where CE2 enzymes have been noted to have a potential catalytic aspartate residue, there often exists a tryptophan that sits between the catalytic histidine and aspartate residues, thereby preventing the aspartate from completing the triad <cite>Montanier2009</cite>. ''Cj''CE2A ([{{PDBlink}}2WAA PDB ID 2WAA]) is an exception, as it has a functioning catalytic triad with no interrupting tryptophan residue <cite>Montanier2009</cite>. Beyond the catalytic residues, CE2 enzymes have also been noted to possess an aromatic amino acid (either a tyrosine or a tryptophan) above their binding clefts that promotes greater substrate specificity <cite>Montanier2009 Till2013</cite>. Lastly, the oxyanion hole is comprised of backbone atoms from the catalytic serine, a glycine, and an asparagine residue that are invariant across the CE2 family and commonly found in other related acetyl-esterases <cite>Montanier2009 Till2013</cite>. | |
| − | Most CE2 family members contain a catalytic dyad | ||
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | [[File:CJCE2B_cropped.png||thumb|300px|right|'''Figure 1.''' ''Cj''CE2B from ''C. | + | [[File:CJCE2B_cropped.png||thumb|300px|right|'''Figure 1.''' ''Cj''CE2B from ''C. japonicus'' ([{{PDBlink}}2W9X PDB ID 2W9X]) ]] |
| − | The possession of an α/β hydrolase fold containing | + | The possession of an α/β hydrolase fold containing a catalytic serine nucleophile suggests that the reaction mechanism may proceed similar to other enzymes in the SGNH family. An example of a proposed reaction mechanism associated with the SGNH family of enzymes involves a catalytic histidine residue acting as a general base. The histidine abstracts a proton from the hydroxyl group of the catalytic serine, thereby rendering it nucleophilic. The serine can then attack the ester bond of the substrate and lead to the formation of a serine-substrate tetrahedral intermediate that is stabilized by the residues of the enzyme's oxyanion hole. The histidine then acts as a general acid and donates a proton to the sugar substrate that leads to its release while the acetyl group remains attached to the catalytic serine. The histidine then acts as a general base and deprotonates a water molecule, so that it can attack the acetyl-serine ester linkage, thereby generating a new tetrahedral intermediate that is also stabilized by the residues of the oxyanion hole. Upon collapse of this transition state, the acetyl group is released from the enzyme and the serine is re-protonated so that it is ready for another catalytic cycle <cite>Alalouf2011</cite>. |
| − | The characterized enzymes were all tested using ''p''NP-Ac, | + | The characterized enzymes were all tested using ''p''NP-Ac as a substrate, with ''k''<sub>cat</sub>/''K''<sub>M</sub> values of 2.01, 0.71, 0.38 and 3.13 s<sup>-1</sup>µM<sup>-1</sup> for Est2A ([{{PDBlink}}3U37 PDB ID 3U37]), ''Ct''CE2 ([{{PDBlink}}2WAO PDB ID 2WAO]), ''Cj''CE2A ([{{PDBlink}}2WAA PDB ID 2WAA]), and ''Cj''CE2B ([{{PDBlink}}2W9X PDB ID 2W9X]), respectively. Est2A was also tested using ''p''-nitrophenyl butyrate that resulted in a ''k''<sub>cat</sub>/''K''<sub>M</sub> value of 2.33 x 10<sup>-3</sup> s<sup>-1</sup>µM<sup>-1</sup> showing the significant decrease in catalytic efficiency as substrate size increased <cite>Till2013</cite>. In order to test for positional specificity, the enzyme kinetics of ''Ct''CE2, ''Cj''CE2B, and ''Cj''CE2C were tested using 2-, 3-, and 4-''O''-acetyl-nitrophenyl-β-D-xylopyranosides that showed increased ''k''<sub>cat</sub>/''K''<sub>M</sub> values for the hydrolysis of the substrate at position 3 and 4 over position 2 <cite>Topakas2010</cite>. Enzyme kinetic assays on birchwood xylan showed ''k''<sub>cat</sub>/''K''<sub>M</sub> values of 7.33 x 10<sup>-5</sup>, 7.67 x 10<sup>-4</sup>, and 2.33 x 10<sup>-4</sup> s<sup>-1</sup>µM<sup>-1</sup> for ''Ct''CE2, ''Cj''CE2A, and ''Cj''CE2B, respectively. When the assay was performed with glucomannan as the substrate for these enzymes, the ''k''<sub>cat</sub>/''K''<sub>M</sub> values were 9.67 x 10<sup>-4</sup>, 6.5 x 10<sup>-4</sup>, and 2.68 x 10<sup>-2</sup> s<sup>-1</sup>µM<sup>-1</sup> for ''Ct''CE2, ''Cj''CE2A and ''Cj''CE2B, respectively; thereby suggesting a substrate preference for glucomannan among CE2 family members <cite>Montanier2009</cite> |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| + | All reported CE2 structures are α/β-hydrolases, e.g. ''Clostridium thermocellum''’s ''Ct''CE2 ([{{PDBlink}}2WAO PDB ID 2WAO]), ''Cellvibrio japonicus''’ ''Cj''CE2A ([{{PDBlink}}2WAA PDB ID 2WAA]) and ''Cj''CE2B ([{{PDBlink}}2W9X PDB ID 2W9X])(See Fig. 1), and ''Butyrivibrio proteoclasticus''’ Est2A ([{{PDBlink}}3U37 PDB ID 3U37]). They contain an N-terminal β-sheet “jelly-roll” domain that acts as a carbohydrate binding domain (CBM) and is linked to a C-terminal domain that contains the α/β-hydrolase fold (SGNH-hydrolase motif) <cite>Montanier2009 Till2013</cite>. The common structure of the N-terminal β-sheet “jelly-roll” domain across CE2 enzymes is comprised of two opposing β-sheets that have 4 and 5 β-strands, respectively <cite>Till2013</cite>. The α/β-hydrolase domain that is C-terminal to the jelly roll consists of a three layered α/β stack composed of five β-strands, arranged in parallel to form a central β-sheet, that is packed between α-helicies. In the case of ''Ct''CE2 ([{{PDBlink}}2WAO PDB ID 2WAO]), ''Cj''CE2A ([{{PDBlink}}2WAA PDB ID 2WAA]), and ''Cj''CE2B ([{{PDBlink}}2W9X PDB ID 2W9X]), the sheet has 5 α-helices in total packed on each side <cite>Montanier2009</cite>, but Est2A has 9 α-helices packing both sides of its β-sheet. | ||
| − | + | The CE2 family members are typically monomeric, but there are some exceptions. Specifically, Est2A has been found to form tetramers that combine to make an overall octameric structure <cite>Montanier2009 Till2013</cite>. The overall structure of ''Ct''CE2 ([{{PDBlink}}2WAO PDB ID 2WAO]) is also unique because this domain is connected to the C-terminal end of a GH5 family cellulase protein, ''Ct''Cel5C ([{{PDBlink}}4IM4 PDB ID 4IM4]), that make up a modular protein, called ''Ct''Cel5C-CE2. This protein is incorporated into cell-wall degrading cellulosomes in ''C. thermocellum'' <cite>Montanier2009 Bayer2004</cite>. | |
| − | |||
| − | |||
| − | |||
| − | The CE2 family members are typically monomeric, but there are some exceptions. Specifically, Est2A has been found to form tetramers that combine to make an overall octameric structure <cite>Montanier2009 Till2013</cite>. The overall structure of ''Ct''CE2 also | ||
== Family Firsts == | == Family Firsts == | ||
;First characterized: | ;First characterized: | ||
| − | The first instance of CE2 family characterization was from the investigation of the BnaA, BnaB, and BnaC proteins that were discovered via cDNA library sequencing of Neocallimastix patriciarum <cite>Dalrymple1997</cite>. BnaA and BnaC proteins exhibited acetyl xylan esterase ability. BnaB exhibited high sequence similarity to an uncharacterized C-terminal domain | + | The first instance of CE2 family characterization was from the investigation of the BnaA, BnaB, and BnaC proteins that were discovered via cDNA library sequencing of ''Neocallimastix patriciarum'' <cite>Dalrymple1997</cite>. BnaA and BnaC proteins exhibited acetyl xylan esterase ability. BnaB exhibited high sequence similarity to an uncharacterized C-terminal domain found in CelE <cite>Hall1988</cite>. The uncharacterized domain of CelE later became known as ''Ct''CE2 ([{{PDBlink}}2WAO PDB ID 2WAO])) <cite>Montanier2009</cite>. |
;First mechanistic insight: | ;First mechanistic insight: | ||
| − | The catalytic dyad of Ser-His residues was confirmed by the arrangement of these residues in the crystal structures of ''Ct''CE2 ([ | + | The catalytic dyad of Ser-His residues was confirmed by the arrangement of these residues in the crystal structures of ''Ct''CE2 ([{{PDBlink}}2WAO PDB ID 2WAO]), ''Cj''CE2A ([{{PDBlink}}2WAA PDB ID 2WAA]), and ''Cj''CE2B ([{{PDBlink}}2W9X PDB ID 2W9X]) as well as sequence alignment analysis showing its invariance across the CE2 family <cite>Montanier2009</cite>. |
;First 3-D structure: | ;First 3-D structure: | ||
| − | The first 3-D structures of CE2 family members, ''Ct''CE2, ''Cj''CE2A and ''Cj''CE2B were revealed in the same study that confirmed the catalytic mechanism of the family <cite>Montanier2009</cite>. | + | The first 3-D structures of CE2 family members, ''Ct''CE2 ([{{PDBlink}}2WAA PDB ID 2WAA]), ''Cj''CE2A ([{{PDBlink}}2WAA PDB ID 2WAA]) and ''Cj''CE2B ([{{PDBlink}}2W9X PDB ID 2W9X]) were revealed in the same study that confirmed the catalytic mechanism of the family <cite>Montanier2009</cite>. |
== References == | == References == | ||
Latest revision as of 13:18, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Carbohydrate Esterase Family CE2 | |
| Fold | α/β-hydrolase |
| Mechanism | Serine Hydrolase |
| Active site residues | Known, Catalytic Dyad |
| CAZy DB link | |
| https://www.cazy.org/CE2.html | |
Substrate specificities
All of the well characterized carbohydrate esterase family 2 enzymes have been shown to remove acetate groups from the synthetic molecule, 4-nitrophenyl acetate (pNP-Ac) [1, 2]. CE2 family members have also demonstrated preferential de-O-acetylation of xylopyranosides at positions 3 and 4, over the 2 position. In expanded substrate profiles, CE2 enzymes were also noted to deacetylate glucopyranosyl and mannopyranosyl residues at the 6-O position. The greater catalytic activity when deacetylating mannopyranosyl and glucopyranosyl compared to xylopyranosides has prompted the naming of some CE2 family members as 6-de-O-acetylases [3].
Catalytic Residues
Most CE2 family members contain a catalytic dyad [1]. For example, the structurally characterized CtCE2 (PDB ID 2WAO) from Clostridium thermocellum, CjCE2A (PDB ID 2WAA) from Cellvibrio japonicus, and Est2A (PDB ID 3U37) from Butyrivibrio proteoclasticus contain conserved serine and histidine residues that form the catalytic dyad and lack a third aspartate residue that is typically found in serine esterase triads [1, 2]. Without the aspartate residue, the histidine of the catalytic dyads are supported by main-chain carbonyl groups provided by a backbone amino acid. In cases where CE2 enzymes have been noted to have a potential catalytic aspartate residue, there often exists a tryptophan that sits between the catalytic histidine and aspartate residues, thereby preventing the aspartate from completing the triad [1]. CjCE2A (PDB ID 2WAA) is an exception, as it has a functioning catalytic triad with no interrupting tryptophan residue [1]. Beyond the catalytic residues, CE2 enzymes have also been noted to possess an aromatic amino acid (either a tyrosine or a tryptophan) above their binding clefts that promotes greater substrate specificity [1, 2]. Lastly, the oxyanion hole is comprised of backbone atoms from the catalytic serine, a glycine, and an asparagine residue that are invariant across the CE2 family and commonly found in other related acetyl-esterases [1, 2].
Kinetics and Mechanism
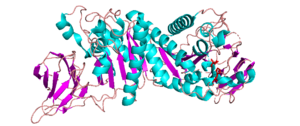
The possession of an α/β hydrolase fold containing a catalytic serine nucleophile suggests that the reaction mechanism may proceed similar to other enzymes in the SGNH family. An example of a proposed reaction mechanism associated with the SGNH family of enzymes involves a catalytic histidine residue acting as a general base. The histidine abstracts a proton from the hydroxyl group of the catalytic serine, thereby rendering it nucleophilic. The serine can then attack the ester bond of the substrate and lead to the formation of a serine-substrate tetrahedral intermediate that is stabilized by the residues of the enzyme's oxyanion hole. The histidine then acts as a general acid and donates a proton to the sugar substrate that leads to its release while the acetyl group remains attached to the catalytic serine. The histidine then acts as a general base and deprotonates a water molecule, so that it can attack the acetyl-serine ester linkage, thereby generating a new tetrahedral intermediate that is also stabilized by the residues of the oxyanion hole. Upon collapse of this transition state, the acetyl group is released from the enzyme and the serine is re-protonated so that it is ready for another catalytic cycle [4].
The characterized enzymes were all tested using pNP-Ac as a substrate, with kcat/KM values of 2.01, 0.71, 0.38 and 3.13 s-1µM-1 for Est2A (PDB ID 3U37), CtCE2 (PDB ID 2WAO), CjCE2A (PDB ID 2WAA), and CjCE2B (PDB ID 2W9X), respectively. Est2A was also tested using p-nitrophenyl butyrate that resulted in a kcat/KM value of 2.33 x 10-3 s-1µM-1 showing the significant decrease in catalytic efficiency as substrate size increased [2]. In order to test for positional specificity, the enzyme kinetics of CtCE2, CjCE2B, and CjCE2C were tested using 2-, 3-, and 4-O-acetyl-nitrophenyl-β-D-xylopyranosides that showed increased kcat/KM values for the hydrolysis of the substrate at position 3 and 4 over position 2 [3]. Enzyme kinetic assays on birchwood xylan showed kcat/KM values of 7.33 x 10-5, 7.67 x 10-4, and 2.33 x 10-4 s-1µM-1 for CtCE2, CjCE2A, and CjCE2B, respectively. When the assay was performed with glucomannan as the substrate for these enzymes, the kcat/KM values were 9.67 x 10-4, 6.5 x 10-4, and 2.68 x 10-2 s-1µM-1 for CtCE2, CjCE2A and CjCE2B, respectively; thereby suggesting a substrate preference for glucomannan among CE2 family members [1]
Three-dimensional structures
All reported CE2 structures are α/β-hydrolases, e.g. Clostridium thermocellum’s CtCE2 (PDB ID 2WAO), Cellvibrio japonicus’ CjCE2A (PDB ID 2WAA) and CjCE2B (PDB ID 2W9X)(See Fig. 1), and Butyrivibrio proteoclasticus’ Est2A (PDB ID 3U37). They contain an N-terminal β-sheet “jelly-roll” domain that acts as a carbohydrate binding domain (CBM) and is linked to a C-terminal domain that contains the α/β-hydrolase fold (SGNH-hydrolase motif) [1, 2]. The common structure of the N-terminal β-sheet “jelly-roll” domain across CE2 enzymes is comprised of two opposing β-sheets that have 4 and 5 β-strands, respectively [2]. The α/β-hydrolase domain that is C-terminal to the jelly roll consists of a three layered α/β stack composed of five β-strands, arranged in parallel to form a central β-sheet, that is packed between α-helicies. In the case of CtCE2 (PDB ID 2WAO), CjCE2A (PDB ID 2WAA), and CjCE2B (PDB ID 2W9X), the sheet has 5 α-helices in total packed on each side [1], but Est2A has 9 α-helices packing both sides of its β-sheet.
The CE2 family members are typically monomeric, but there are some exceptions. Specifically, Est2A has been found to form tetramers that combine to make an overall octameric structure [1, 2]. The overall structure of CtCE2 (PDB ID 2WAO) is also unique because this domain is connected to the C-terminal end of a GH5 family cellulase protein, CtCel5C (PDB ID 4IM4), that make up a modular protein, called CtCel5C-CE2. This protein is incorporated into cell-wall degrading cellulosomes in C. thermocellum [1, 5].
Family Firsts
- First characterized
The first instance of CE2 family characterization was from the investigation of the BnaA, BnaB, and BnaC proteins that were discovered via cDNA library sequencing of Neocallimastix patriciarum [6]. BnaA and BnaC proteins exhibited acetyl xylan esterase ability. BnaB exhibited high sequence similarity to an uncharacterized C-terminal domain found in CelE [7]. The uncharacterized domain of CelE later became known as CtCE2 (PDB ID 2WAO)) [1].
- First mechanistic insight
The catalytic dyad of Ser-His residues was confirmed by the arrangement of these residues in the crystal structures of CtCE2 (PDB ID 2WAO), CjCE2A (PDB ID 2WAA), and CjCE2B (PDB ID 2W9X) as well as sequence alignment analysis showing its invariance across the CE2 family [1].
- First 3-D structure
The first 3-D structures of CE2 family members, CtCE2 (PDB ID 2WAA), CjCE2A (PDB ID 2WAA) and CjCE2B (PDB ID 2W9X) were revealed in the same study that confirmed the catalytic mechanism of the family [1].
References
- Montanier C, Money VA, Pires VM, Flint JE, Pinheiro BA, Goyal A, Prates JA, Izumi A, Stålbrand H, Morland C, Cartmell A, Kolenova K, Topakas E, Dodson EJ, Bolam DN, Davies GJ, Fontes CM, and Gilbert HJ. (2009). The active site of a carbohydrate esterase displays divergent catalytic and noncatalytic binding functions. PLoS Biol. 2009;7(3):e71. DOI:10.1371/journal.pbio.1000071 |
- Till M, Goldstone DC, Attwood GT, Moon CD, Kelly WJ, and Arcus VL. (2013). Structure and function of an acetyl xylan esterase (Est2A) from the rumen bacterium Butyrivibrio proteoclasticus. Proteins. 2013;81(5):911-7. DOI:10.1002/prot.24254 |
- Topakas E, Kyriakopoulos S, Biely P, Hirsch J, Vafiadi C, and Christakopoulos P. (2010). Carbohydrate esterases of family 2 are 6-O-deacetylases. FEBS Lett. 2010;584(3):543-8. DOI:10.1016/j.febslet.2009.11.095 |
- Alalouf O, Balazs Y, Volkinshtein M, Grimpel Y, Shoham G, and Shoham Y. (2011). A new family of carbohydrate esterases is represented by a GDSL hydrolase/acetylxylan esterase from Geobacillus stearothermophilus. J Biol Chem. 2011;286(49):41993-42001. DOI:10.1074/jbc.M111.301051 |
- Bayer EA, Belaich JP, Shoham Y, and Lamed R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521-54. DOI:10.1146/annurev.micro.57.030502.091022 |
- Dalrymple BP, Cybinski DH, Layton I, McSweeney CS, Xue GP, Swadling YJ, and Lowry JB. (1997). Three Neocallimastix patriciarum esterases associated with the degradation of complex polysaccharides are members of a new family of hydrolases. Microbiology (Reading). 1997;143 ( Pt 8):2605-2614. DOI:10.1099/00221287-143-8-2605 |
- Hall J, Hazlewood GP, Barker PJ, and Gilbert HJ. (1988). Conserved reiterated domains in Clostridium thermocellum endoglucanases are not essential for catalytic activity. Gene. 1988;69(1):29-38. DOI:10.1016/0378-1119(88)90375-7 |