CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 91"
Daichi Ito (talk | contribs) |
|||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
* [[Author]]: [[User:Daichi Ito|Daichi Ito]] | * [[Author]]: [[User:Daichi Ito|Daichi Ito]] | ||
* [[Responsible Curator]]: [[User:Elizabeth Ficko-Blean|Elizabeth Ficko-Blean]] | * [[Responsible Curator]]: [[User:Elizabeth Ficko-Blean|Elizabeth Ficko-Blean]] | ||
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | CBM91 from ''Paenibacillus xynaniclasticus'' bound to oat spelt xylan with ''K''<sub>a</sub> value of 2.0×10<sup>-5</sup> M<sup>-1</sup>, and bound birchwood xylan. | + | ''Px''CBM91 from ''Paenibacillus xynaniclasticus'' bound to oat spelt xylan with ''K''<sub>a</sub> value of 2.0×10<sup>-5</sup> M<sup>-1</sup>, and bound birchwood xylan <cite>Ito2022</cite>. It did not bind to lichenan or the cellulosic substrates carboxymethyl-cellulose or ball-milled cellulose <cite>Ito2022</cite>. Therefore, ''Px''CBM91 can recognize and bind to insoluble xylan <cite>Ito2022</cite>. |
== Structural Features == | == Structural Features == | ||
| − | [[Image:The_structure_of_PxXyl43A.png|thumb|300px|right|'''Figure 1. The structure of ''Px''Xyl43A and CBM91. | + | [[Image:The_structure_of_PxXyl43A.png|thumb|300px|right|'''Figure 1. The structure of ''Px''Xyl43A and ''Px''CBM91 <cite>Ito2023</cite>. |
| − | '''The prediction | + | '''The structure prediction by Alpha Fold 2 of ''Px''CBM91 (red). This CBM91 is appended to the catalytic domain of ''Px''Xyl43A (green).]] |
| − | Alpha Fold 2 | + | Alpha Fold 2 structural analysis of ''Px''CBM91 exhibited a β-sandwich fold consisted of 12 β-strands and two opposing antiparallel beta sheets <cite>Ito2023</cite>. The concave surface and loops around it connecting the β-strands possess several hydrophobic amino acid residues, the surface is expected to be the binding site <cite>Ito2023</cite>. Another CBM91 study suggests the CBM91 may contribute a small loop to extend the enzyme active site <cite>Pang2024</cite>. |
| + | |||
== Functionalities == | == Functionalities == | ||
| − | + | ''Paenibacillus xylaniclastuicus'' was isolated from an anaerobic digester fed with pineapple waste and grows with xylose as sole carbon source <cite>Tachaapaikoon2012, Ratanakhanockchai2012</cite>. ''P. xylaniclastuicus'' likely degrades xylosic substrates efficiently because it has a lot of genes encoding xylolytic enzymes. Xylosidases, like ''Px''Xyl43A, which produces xylose from xylan and xylooligosaccharides is one of the vital enzymes. Therefore, the appended ''Px''CBM91 contributes to producing the carbon sources for the growth of this xylolytic bacteria. | |
| − | Paenibacillus xylaniclastuicus was isolated from anaerobic digester fed with pineapple waste and grows with xylose as sole carbon source. | ||
| − | Xylosidases, like | ||
| + | CBM91 are often connected to β-xylosidases belonging to glycoside hydrolase family 43 ([[GH43]]) (see [http://www.cazy.org/CBM91_structure.html CBM91 page of the CAZy Database]). CBM91 binding to the substrates would place the catalytic domain in the vicinity of substrates in which substrate concentration is high. These enzymes would utilize CBM91 as a tool for efficient saccharification in combination with other xylanases which release xylobiose and/or xylo-oligosaccharides from insoluble substrates. | ||
| + | In the ''Physcomitrellae patens'' β-xylosidase/α-L-arabinofuranosidase bifunctional [[GH43]] enzyme the CBM91 at its C-terminus was essential for catalytic activity on artificial substrates and the deletion mutant greatly reduced activity on natural substrates <cite>Pang2024</cite>. | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First Identified:''Px''CBM91 from ''Px''Xyl43A of ''Paenibacillus xynaniclasticus'' strain TW1 <cite>Ito2022</cite>. | + | ;First Identified: Xylan binding was first identified in ''Px''CBM91 from ''Px''Xyl43A of ''Paenibacillus xynaniclasticus'' strain TW1 <cite>Ito2022</cite>. |
| − | ;First Structural Characterization: β-D-xylosidase, a family 43 glycoside hydrolase from ''Clostridium acetobutylicum'' ATCC 824 [{{PDBlink}}1Y7B PDB ID 1Y7B]. | + | ;First Structural Characterization: β-D-xylosidase, a family 43 glycoside hydrolase from ''Clostridium acetobutylicum'' ATCC 824 (Released: 2005-01-25) [{{PDBlink}}1Y7B PDB ID 1Y7B] <cite>StructuralGenomics</cite>. |
== References == | == References == | ||
<biblio> | <biblio> | ||
| + | #Tachaapaikoon2012 C. Tachaapaikoon, S. Tanasupawat, P. Pason, S. Sornyotha, R. Waeonukul, K.L. Kyu and K. Ratanakhanockchai: ''Paenibacillus xylaniclasticus'' sp. nov., a xylanolytic-cellulolytic bacterium isolated from sludge in an anaerobic digester. J.Microbiol., 50, 394–400 (2012) [https://doi.org/10.1007/s12275-012-1480-3 DOI:10.1007/s12275-012-1480-3] | ||
#Ito2022 pmid=36312872 | #Ito2022 pmid=36312872 | ||
| − | + | #Ito2023 Ito, D., 2023. Characterization of plant cell wall degrading enzymes from Paenibacillus sp., 2023, Mie University, Ph. D. thesis. https://dl.ndl.go.jp/pid/12910195/1/1 | |
| − | + | ||
| − | #Ito2023 Ito, D., 2023. Characterization of plant cell wall degrading enzymes from Paenibacillus sp. Ph. D. | + | #Pang2024 pmid=38556222 |
| + | #Ratanakhanockchai2012 K. Ratanakhanockchai, C. Tachaapaikoon, K.L. Kyu and P. Pason: A novel multienzyme complex from a newly isolated facultative anaerobic bacterium, ''Paenibacillus'' sp. TW1. Act. Biol. Hung., 63, 288–300 (2012) [https://doi.org/10.1556/ABiol.63.2012.2.10 DOI:10.1556/ABiol.63.2012.2.10] | ||
| + | #StructuralGenomics Teplyakov, A., Fedorov, E., Gilliland, G.L., Almo, S.C., Burley, S.K., New York SGX Research Center for Structural Genomics (NYSGXRC) | ||
| + | |||
</biblio> | </biblio> | ||
<!-- Do not delete this Category tag --> | <!-- Do not delete this Category tag --> | ||
[[Category:Carbohydrate Binding Module Families|CBM091]] | [[Category:Carbohydrate Binding Module Families|CBM091]] | ||
Latest revision as of 04:55, 29 July 2025
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM91.html |
Ligand specificities
PxCBM91 from Paenibacillus xynaniclasticus bound to oat spelt xylan with Ka value of 2.0×10-5 M-1, and bound birchwood xylan [1]. It did not bind to lichenan or the cellulosic substrates carboxymethyl-cellulose or ball-milled cellulose [1]. Therefore, PxCBM91 can recognize and bind to insoluble xylan [1].
Structural Features
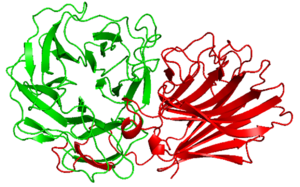
Alpha Fold 2 structural analysis of PxCBM91 exhibited a β-sandwich fold consisted of 12 β-strands and two opposing antiparallel beta sheets [2]. The concave surface and loops around it connecting the β-strands possess several hydrophobic amino acid residues, the surface is expected to be the binding site [2]. Another CBM91 study suggests the CBM91 may contribute a small loop to extend the enzyme active site [3].
Functionalities
Paenibacillus xylaniclastuicus was isolated from an anaerobic digester fed with pineapple waste and grows with xylose as sole carbon source [4, 5]. P. xylaniclastuicus likely degrades xylosic substrates efficiently because it has a lot of genes encoding xylolytic enzymes. Xylosidases, like PxXyl43A, which produces xylose from xylan and xylooligosaccharides is one of the vital enzymes. Therefore, the appended PxCBM91 contributes to producing the carbon sources for the growth of this xylolytic bacteria.
CBM91 are often connected to β-xylosidases belonging to glycoside hydrolase family 43 (GH43) (see CBM91 page of the CAZy Database). CBM91 binding to the substrates would place the catalytic domain in the vicinity of substrates in which substrate concentration is high. These enzymes would utilize CBM91 as a tool for efficient saccharification in combination with other xylanases which release xylobiose and/or xylo-oligosaccharides from insoluble substrates.
In the Physcomitrellae patens β-xylosidase/α-L-arabinofuranosidase bifunctional GH43 enzyme the CBM91 at its C-terminus was essential for catalytic activity on artificial substrates and the deletion mutant greatly reduced activity on natural substrates [3].
Family Firsts
- First Identified
- Xylan binding was first identified in PxCBM91 from PxXyl43A of Paenibacillus xynaniclasticus strain TW1 [1].
- First Structural Characterization
- β-D-xylosidase, a family 43 glycoside hydrolase from Clostridium acetobutylicum ATCC 824 (Released: 2005-01-25) PDB ID 1Y7B [6].
References
- Ito D, Nakano E, Karita S, Umekawa M, Ratanakhanokchai K, and Tachaapaikoon C. (2022). Characterization of a GH Family 43 β-Xylosidase Having a Novel Carbohydrate-binding Module from Paenibacillus xylaniclasticus Strain TW1. J Appl Glycosci (1999). 2022;69(3):65-71. DOI:10.5458/jag.jag.JAG-2022_0001 |
-
Ito, D., 2023. Characterization of plant cell wall degrading enzymes from Paenibacillus sp., 2023, Mie University, Ph. D. thesis. https://dl.ndl.go.jp/pid/12910195/1/1
- Error fetching PMID 38556222:
-
C. Tachaapaikoon, S. Tanasupawat, P. Pason, S. Sornyotha, R. Waeonukul, K.L. Kyu and K. Ratanakhanockchai: Paenibacillus xylaniclasticus sp. nov., a xylanolytic-cellulolytic bacterium isolated from sludge in an anaerobic digester. J.Microbiol., 50, 394–400 (2012) DOI:10.1007/s12275-012-1480-3
-
K. Ratanakhanockchai, C. Tachaapaikoon, K.L. Kyu and P. Pason: A novel multienzyme complex from a newly isolated facultative anaerobic bacterium, Paenibacillus sp. TW1. Act. Biol. Hung., 63, 288–300 (2012) DOI:10.1556/ABiol.63.2012.2.10
-
Teplyakov, A., Fedorov, E., Gilliland, G.L., Almo, S.C., Burley, S.K., New York SGX Research Center for Structural Genomics (NYSGXRC)