CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycosyltransferase Family 138"
| Line 36: | Line 36: | ||
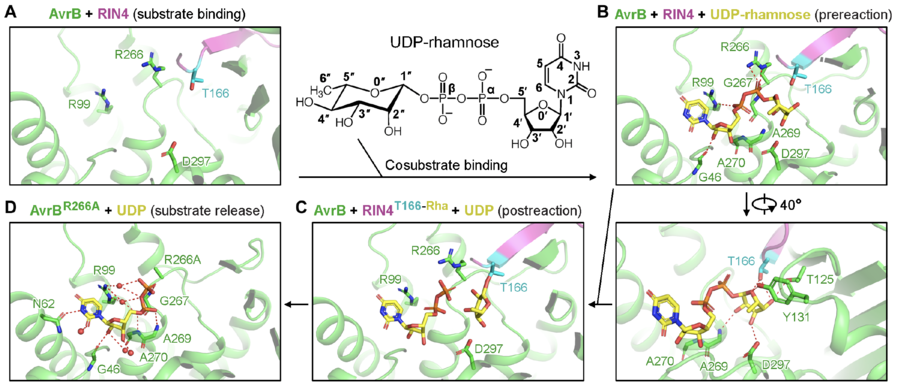
[[File:GT138-figure-2.png|thumb|900px|center|'''Figure 2. Catalysis mechanisms for RIN4 rhamnosylation by AvrB supported by crystal structures.''' ('''A''') AvrB bound with RIN4. ('''B''') UDP-rhamnose bound with AvrB and RIN4. ('''C''') Rhamnose transferred to T166 of RIN4. ('''D''') Release of rhamnosylated RIN4.]] | [[File:GT138-figure-2.png|thumb|900px|center|'''Figure 2. Catalysis mechanisms for RIN4 rhamnosylation by AvrB supported by crystal structures.''' ('''A''') AvrB bound with RIN4. ('''B''') UDP-rhamnose bound with AvrB and RIN4. ('''C''') Rhamnose transferred to T166 of RIN4. ('''D''') Release of rhamnosylated RIN4.]] | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| − | A threonine from the protein substrate directly attack the rhamnose moiety in the co-substrate, UDP-rhamnose. UDP-rhamnose is stabilized by a few residues in the pocket. | + | A threonine (T166) from the protein substrate directly attack the rhamnose moiety in the co-substrate, UDP-rhamnose (Fig. 2). The threonine is close to a histidine and a threonine, which may stabilize the acceptor. UDP-rhamnose is stabilized by a few residues in the pocket. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
Revision as of 20:05, 4 December 2025
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| Glycosyltransferase Family GT138 | |
| Clan | Fido fold |
| Mechanism | Inverting |
| Active site residues | Known |
| CAZy DB link | |
| https://www.cazy.org/GT138.html | |
Substrate specificities
GT138 family of glycosyltransferase is exemplified by AvrB [1]. As a bacterial effector from the plant pathogen Pseudomonas syringae, AvrB utilizes host UDP-rhamnose (or dTDP-rhamnose in vitro) as a co-substrate to modify the host protein RIN4 and causes the programmed cell death (namely hypersensitive response) [1, 2].
Kinetics and Mechanism
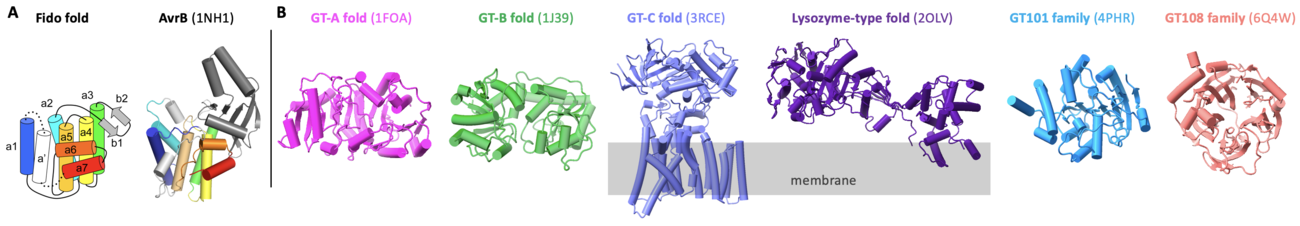
AvrB contains a Fido domain [3, 4] (Fig. 1A), different from other known glycosyltransferases containing folds of GT-A, GT-B, GT-C, lysozyme-type, GT101, and GT108 [5, 6, 7, 8] (Fig. 1B). Interestingly, Fido proteins can also be enzymes with activities of AMPylation [9], phosphorylation [10], UMPylation [11], and phosphocholination [12, 13]. Hence, AvrB is a unique Fido protein that functions as a glycosyltransferase.
The rhamnosylation reaction catalyzed by AvrB does not require divalent cations (e.g., Mg2+). In the reaction, rhamnose is directly transferred to the side chain of a threonine (Fig. 2).
Catalytic Residues
A threonine (T166) from the protein substrate directly attack the rhamnose moiety in the co-substrate, UDP-rhamnose (Fig. 2). The threonine is close to a histidine and a threonine, which may stabilize the acceptor. UDP-rhamnose is stabilized by a few residues in the pocket.
Three-dimensional structures
AvrB represents the prototype for glycosyltransferases of Fido fold. AvrB contains a large internal domain between helix α2 and helix α3 (Fig. 1A). AvrB shares similar structural features with other Fido proteins despite the primary sequences are divergent.
Family Firsts
- First stereochemistry determination
- Content is to be added here.
- First catalytic nucleophile identification
- Content is to be added here.
- First general acid/base residue identification
- Content is to be added here.
- First 3-D structure
- The first structure of GT138 family (Fido type) is the crystal structure of AvrB [3].
References
Error fetching PMID 19503829:
Error fetching PMID 19039103:
Error fetching PMID 24141193:
Error fetching PMID 22504181:
Error fetching PMID 21822290:
Error fetching PMID 23572077:
Error fetching PMID 35536922:
Error fetching PMID 18518825:
Error fetching PMID 25023666:
Error fetching PMID 31513773:
Error fetching PMID 11955429:
Error fetching PMID 15016364:
- Error fetching PMID 38354245:
- Error fetching PMID 11955429:
- Error fetching PMID 15016364:
- Error fetching PMID 19503829:
- Error fetching PMID 35536922:
- Error fetching PMID 18518825:
- Error fetching PMID 25023666:
- Error fetching PMID 31513773:
- Error fetching PMID 19039103:
- Error fetching PMID 24141193:
- Error fetching PMID 22504181:
- Error fetching PMID 21822290:
- Error fetching PMID 23572077: