CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 64"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (50 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | {{CuratorApproved}} | |
| − | + | * [[Author]]: [[User:Julie Grondin|Julie Grondin]] | |
| − | * [[Author]]: | + | * [[Responsible Curator]]: [[User:Harry Brumer|Harry Brumer]] |
| − | * [[Responsible Curator]]: | ||
---- | ---- | ||
| Line 12: | Line 11: | ||
|- | |- | ||
|'''Clan''' | |'''Clan''' | ||
| − | | | + | |none |
|- | |- | ||
|'''Mechanism''' | |'''Mechanism''' | ||
| − | | | + | |inverting |
|- | |- | ||
|'''Active site residues''' | |'''Active site residues''' | ||
| − | | | + | |known |
|- | |- | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
| Line 29: | Line 28: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | + | All characterized members of the GH64 family are laminaripentaose-producing β(1,3)-glucanases ([{{EClink}}3.2.1.39 EC 3.2.1.39]) from the GH64-TLP (thaumatin-like protein) superfamily. They are found in bacteria and fungal species, and are particularly abundant in the genomes of various ''Streptomyces'' and ''Fusarium'' species. Activity has been shown on insoluble and soluble β-1,3-glucans, including curdlan <cite>Nakabayashi1998, Palumbo2003, Wu2009</cite>, colloidal pachyman <cite>Nakabayashi1998, Nishimura2001, Wu2009</cite>, laminarin <cite>Nakabayashi1998, Nishimura2001, Wu2009</cite>, and zymosan A <cite>Nakabayashi1998, Palumbo2003</cite>, a commercial preparation of partially-purified yeast cell walls (contains branched glucans). | |
| − | |||
| − | |||
| − | |||
| − | |||
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | + | GH64 enzymes follow an [[inverting]] mechanism, first shown by <sup>1</sup>H-NMR during the hydrolysis of laminarin <cite>Nishimura2001</cite>, thus operating by a [[Glycoside_hydrolases#Inverting_glycoside_hydrolases|single-displacement mechanism]]. | |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | + | The general acid and base for this family were first proposed in 2009, based on the active site topology and residue orientation observed in the first published structure in this family, of the LPHase from ''Streptoymces matensis'' <cite>Wu2009</cite>. Subsequent enzymological characterization of this enzyme by the same group confirmed the identity of the general acid (Glu154) and general base (Asp170), via site-directed mutagenesis, chemical rescue, and kinetic analysis <cite>Shrestha2011</cite>. | |
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | + | [[File:5H9Y_Lam6.png|400px|thumb|right|'''Figure 1. Structure of ''Pb''Bgl64A in complex with two laminarihexaose chains (forest, pale green).''' ([{{PDBlink}}5H9Y PDB ID 5H9Y]) The barrel domain is shown in light pink, the α/β domain in light blue, and the CBM56 in wheat. The general acid (here, as E236A) and general base (D252) are shown as indigo sticks.]] | |
| + | Each three-dimensional GH64 structure characterized to date shares a crescent-like fold comprising of a barrel domain, and a mixed α/β domain ('''Figure 1'''). This fold is unique among GHs; GH64s are not classified into any existing GH clans. In some instances, the GH64 may also contain a N-terminal [[CBM13]] or [[CBM56]], the latter of which has been shown to bind β-1,3-glucans <cite>Qin2017</cite>. | ||
| + | |||
| + | The interface between the two domains forms a wide, electronegatively-charged groove, which contains the active site. Specifically, the proton donor and base are located in the center of this groove, on strands β7 and β9 of the barrel domain, approximately 7.5 Å apart. The laminaritetraose-bound structure of LPHase from ''Streptoymces matensis'' ([{{PDBlink}}3GD9 PDB ID 3GD9]) reveals that the barrel domain mediates most of the interactions between the sugar and the protein <cite>Wu2009</cite> (NB: In this case, LPHase was co-crystallized with laminaripentaose, however, density was only observed for four glycosyl residues). To date, no significant conformational changes have been noted between ''apo'' and complex structures. | ||
| + | |||
| + | The three-dimensional structure of the catalytically-inactive Blg64A from ''Paenibacillus barengoltzii'', in complex with two laminarihexaose chains ([{{PDBlink}}5H9Y PDB ID 5H9Y]), provides the first structure-based evidence to explain how GH64s enzymes might bind to the triple-helical structure adopted by β-1,3-glucans ''in vivo'' <cite>Qin2017</cite> ('''Figure 1'''). In this structure, the 40 Å-long groove containing the active site is 15 Å wide, and thus accommodates the two twisted laminarihexaose chains. The surface of the groove is also sufficiently large to accommodate the triple helix of linear β-1,3-glucans such as curdlan, however, the β-1,6 branching typical of laminarin would be sterically hindered, explaining the lower activity of this enzyme on laminarin. To date, there is no experimental evidence to explain how the triple helix is hydrolyzed by GH64s. | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First stereochemistry determination: | + | ;First stereochemistry determination: Laminaripentaose-producing β-1,3-glucanase (LPHase) from ''Streptomyces matensis'' DIC-108 by <sup>1</sup>H-NMR <cite>Nishimura2001</cite>. |
| − | ;First catalytic nucleophile identification: | + | ;First catalytic nucleophile identification: Laminaripentaose-producing β-1,3-glucanase (LPHase) from ''Streptomyces matensis'' DIC-108 by site-directed mutagenesis, chemical rescue, and kinetic analysis <cite>Shrestha2011</cite>. |
| − | ;First general acid/base residue identification: | + | ;First general acid/base residue identification: Laminaripentaose-producing β-1,3-glucanase (LPHase) from ''Streptomyces matensis'' DIC-108 by site-directed mutagenesis, chemical rescue, and kinetic analysis <cite>Shrestha2011</cite>. |
| − | ;First 3-D structure: | + | ;First 3-D structure: Laminaripentaose-producing β-1,3-glucanase (LPHase) from ''Streptomyces matensis'' DIC-108 by X-ray crystallography.<cite>Wu2009</cite> |
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Nakabayashi1998 Nakabayashi M, Nishijima T, Ehara G, Nikaidou N, Nishihashi H, and Watanabe T. (1998) Structure of the gene encoding laminaripentaose-producing β-1,3-glucanase (LPHase) of Streptomyces matensis DIC-108. ''J. Ferment. Bioengineer.'' '''85''', 459-464. [http://dx.doi.org/10.1016/s0922-338x(98)80062-7 DOI:10.1016/s0922-338x(98)80062-7] |
| − | + | #Palumbo2003 pmid=12867444 | |
| + | #Wu2009 pmid=19640850 | ||
| + | #Nishimura2001 pmid=11418137 | ||
| + | #Shrestha2011 pmid=21705773 | ||
| + | #Qin2017 pmid=28787048 | ||
</biblio> | </biblio> | ||
[[Category:Glycoside Hydrolase Families|GH064]] | [[Category:Glycoside Hydrolase Families|GH064]] | ||
Latest revision as of 13:19, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH64 | |
| Clan | none |
| Mechanism | inverting |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH64.html | |
Substrate specificities
All characterized members of the GH64 family are laminaripentaose-producing β(1,3)-glucanases (EC 3.2.1.39) from the GH64-TLP (thaumatin-like protein) superfamily. They are found in bacteria and fungal species, and are particularly abundant in the genomes of various Streptomyces and Fusarium species. Activity has been shown on insoluble and soluble β-1,3-glucans, including curdlan [1, 2, 3], colloidal pachyman [1, 3, 4], laminarin [1, 3, 4], and zymosan A [1, 2], a commercial preparation of partially-purified yeast cell walls (contains branched glucans).
Kinetics and Mechanism
GH64 enzymes follow an inverting mechanism, first shown by 1H-NMR during the hydrolysis of laminarin [4], thus operating by a single-displacement mechanism.
Catalytic Residues
The general acid and base for this family were first proposed in 2009, based on the active site topology and residue orientation observed in the first published structure in this family, of the LPHase from Streptoymces matensis [3]. Subsequent enzymological characterization of this enzyme by the same group confirmed the identity of the general acid (Glu154) and general base (Asp170), via site-directed mutagenesis, chemical rescue, and kinetic analysis [5].
Three-dimensional structures
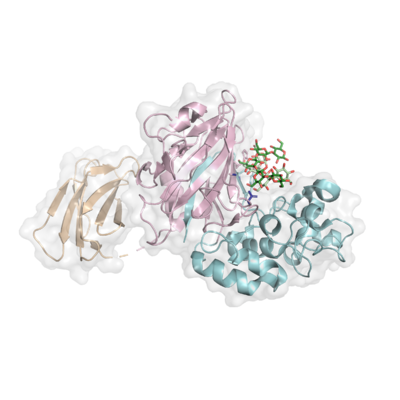
Each three-dimensional GH64 structure characterized to date shares a crescent-like fold comprising of a barrel domain, and a mixed α/β domain (Figure 1). This fold is unique among GHs; GH64s are not classified into any existing GH clans. In some instances, the GH64 may also contain a N-terminal CBM13 or CBM56, the latter of which has been shown to bind β-1,3-glucans [6].
The interface between the two domains forms a wide, electronegatively-charged groove, which contains the active site. Specifically, the proton donor and base are located in the center of this groove, on strands β7 and β9 of the barrel domain, approximately 7.5 Å apart. The laminaritetraose-bound structure of LPHase from Streptoymces matensis (PDB ID 3GD9) reveals that the barrel domain mediates most of the interactions between the sugar and the protein [3] (NB: In this case, LPHase was co-crystallized with laminaripentaose, however, density was only observed for four glycosyl residues). To date, no significant conformational changes have been noted between apo and complex structures.
The three-dimensional structure of the catalytically-inactive Blg64A from Paenibacillus barengoltzii, in complex with two laminarihexaose chains (PDB ID 5H9Y), provides the first structure-based evidence to explain how GH64s enzymes might bind to the triple-helical structure adopted by β-1,3-glucans in vivo [6] (Figure 1). In this structure, the 40 Å-long groove containing the active site is 15 Å wide, and thus accommodates the two twisted laminarihexaose chains. The surface of the groove is also sufficiently large to accommodate the triple helix of linear β-1,3-glucans such as curdlan, however, the β-1,6 branching typical of laminarin would be sterically hindered, explaining the lower activity of this enzyme on laminarin. To date, there is no experimental evidence to explain how the triple helix is hydrolyzed by GH64s.
Family Firsts
- First stereochemistry determination
- Laminaripentaose-producing β-1,3-glucanase (LPHase) from Streptomyces matensis DIC-108 by 1H-NMR [4].
- First catalytic nucleophile identification
- Laminaripentaose-producing β-1,3-glucanase (LPHase) from Streptomyces matensis DIC-108 by site-directed mutagenesis, chemical rescue, and kinetic analysis [5].
- First general acid/base residue identification
- Laminaripentaose-producing β-1,3-glucanase (LPHase) from Streptomyces matensis DIC-108 by site-directed mutagenesis, chemical rescue, and kinetic analysis [5].
- First 3-D structure
- Laminaripentaose-producing β-1,3-glucanase (LPHase) from Streptomyces matensis DIC-108 by X-ray crystallography.[3]
References
-
Nakabayashi M, Nishijima T, Ehara G, Nikaidou N, Nishihashi H, and Watanabe T. (1998) Structure of the gene encoding laminaripentaose-producing β-1,3-glucanase (LPHase) of Streptomyces matensis DIC-108. J. Ferment. Bioengineer. 85, 459-464. DOI:10.1016/s0922-338x(98)80062-7
- Palumbo JD, Sullivan RF, and Kobayashi DY. (2003). Molecular characterization and expression in Escherichia coli of three beta-1,3-glucanase genes from Lysobacter enzymogenes strain N4-7. J Bacteriol. 2003;185(15):4362-70. DOI:10.1128/JB.185.15.4362-4370.2003 |
- Wu HM, Liu SW, Hsu MT, Hung CL, Lai CC, Cheng WC, Wang HJ, Li YK, and Wang WC. (2009). Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase. J Biol Chem. 2009;284(39):26708-15. DOI:10.1074/jbc.M109.010983 |
- Nishimura T, Bignon C, Allouch J, Czjzek M, Darbon H, Watanabe T, and Henrissat B. (2001). Streptomyces matensis laminaripentaose hydrolase is an 'inverting' beta-1,3-glucanase. FEBS Lett. 2001;499(1-2):187-90. DOI:10.1016/s0014-5793(01)02551-0 |
- Shrestha KL, Liu SW, Huang CP, Wu HM, Wang WC, and Li YK. (2011). Characterization and identification of essential residues of the glycoside hydrolase family 64 laminaripentaose-producing-β-1, 3-glucanase. Protein Eng Des Sel. 2011;24(8):617-25. DOI:10.1093/protein/gzr031 |
- Qin Z, Yang D, You X, Liu Y, Hu S, Yan Q, Yang S, and Jiang Z. (2017). The recognition mechanism of triple-helical β-1,3-glucan by a β-1,3-glucanase. Chem Commun (Camb). 2017;53(67):9368-9371. DOI:10.1039/c7cc03330c |