CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 49"
| (19 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:Takashi Tonozuka|Takashi Tonozuka]] and [[User:Takatsugu Miyazaki|Takatsugu Miyazaki]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Takashi Tonozuka|Takashi Tonozuka]] |
---- | ---- | ||
| Line 9: | Line 9: | ||
{| {{Prettytable}} | {| {{Prettytable}} | ||
|- | |- | ||
| − | |{{Hl2}} colspan="2" align="center" |'''Glycoside Hydrolase Family | + | |{{Hl2}} colspan="2" align="center" |'''Glycoside Hydrolase Family GH49''' |
|- | |- | ||
|'''Clan''' | |'''Clan''' | ||
| Line 22: | Line 22: | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
|- | |- | ||
| − | | colspan="2" | | + | | colspan="2" |{{CAZyDBlink}}GH49.html |
|} | |} | ||
</div> | </div> | ||
| Line 29: | Line 29: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | [[ | + | [[File:Fig1_GH49_substrates.png|thumb|600px|right|'''Figure 1. Substrates of GH49 enzymes.''' All enzymes except dextran 1,6-α-isomaltotriosidase are endo-acting glycoside hydrolases. Sulfated arabinan contains sulfate groups at 2- and 3-OH (3S) of L-arabinopyranose residue and possesses side chains of galactose and xylose, which are not shown in this figure.]] |
| + | [[Glycoside hydrolases]] of GH49 cleave α-(1→6)-glucosidic linkages or α-(1→4)-glucosidic linkages of polysaccharides and oligosaccharides containing α-(1→6)-glucosidic linkages, such as dextran and pullulan. The major activities reported for this family of glycoside hydrolases are dextranase (EC [{{EClink}}3.2.1.11 3.2.1.11]), and a dextranase from ''Talaromyces minioluteum'' (formerly known as ''Penicillium minioluteum''), Dex49A, is currently the most characterized enzyme. GH49 dextranases have been found in some bacteria and fungi. Dextran 1,6-α-isomaltotriosidase (EC [{{EClink}}3.2.1.95 3.2.1.95]) from ''Brevibacterium fuscum'' var. ''dextranlyticum'' is an exo-acting enzyme that hydrolyzes dextran from the non-reducing ends to produce isomaltotriose <cite>Mizuno1999</cite>. Isopullulanase (EC [{{EClink}}3.2.1.57 3.2.1.57])from ''Aspergillus brasiliensis'' ATCC 9642 (formerly ''Aspergillus niger'' ATCC 9642) hydrolyzes α-(1→4)-linkages of pullulan to produce isopanose <cite>Sakano1971</cite>. 4-''O''-α-D-Isomaltooligosaccharylmaltooligosaccharide 1,4-α-isomaltooligosaccharohydrolase (IMM-4IH, EC 3.2.1.-) from ''Sarocladium kiliense'' possesses more strict substrate specificity than isopullulanase and cannot hydrolyze pullulan <cite>Kitagawa2023</cite>. As an exception, endo-acting sulfated-arabinan hydrolase (EC 3.2.1-) from ''Phocaeicola plebeius'' degrades sulfated arabinan produced by green algae ''Chaetomorpha'' sp. and ''Cladophora'' sp. <cite>Helbert2019</cite>. | ||
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | Family GH49 α-glycosidases are inverting enzymes, as first shown by NMR on a dextranase Dex49A from '' | + | Family GH49 α-glycosidases are [[inverting]] enzymes, as first shown by NMR on a dextranase Dex49A from ''Talaromyces minioluteum'' <cite>Larsson2003</cite> . Optical rotation analysis of the hydrolysis of panose by isopullulanase supported the inverting mechanism <cite>Akeboshi2004</cite>. |
| + | == Catalytic Residues == | ||
| + | Three Asp residues (Asp376, Asp395, and Asp396 in Dex49A) are conserved in the catalytic center of clan GH-N members, GH49 and [[GH28]] enzymes <cite>Larsson2003 Mizuno2008</cite>, which is also the case for [[GH87]] and [[GH110]]. All three of the Asp mutants of ''A. brasiliensis'' isopullulanase, lost their activities <cite>Akeboshi2004</cite>. The [[general acid]] was first identified in Dex49A from ''Talaromyces minioluteum'' as Asp395 following the three-dimensional structure determination. To date, it is unclear whether either (or both) of the Asp residues (Asp376 and Asp396 in Dex49A) acts as a [[general base]] in the reaction of GH49 and [[GH28]] enzymes <cite>Larsson2003 vanSanten1999 Shimizu2002</cite>. | ||
| + | |||
| + | == Three-dimensional structures == | ||
| + | [[File:Fig2_GH49_structures.png|thumb|800px|center|'''Figure 2. Overall structures of GH49 enzymes.''' (Left to right) ''Aspergillus brasiliensis'' isopullulanase (IPU) in complex with isopanose (PDB ID [{{PDBlink}}2z8g 2Z8G]), ''Talaromyces minioluteum'' dextranase (Dex49A) in complex with isomaltose (PDB ID [{{PDBlink}}1ogm 1OGM]), and ''Arthrobacter oxydans'' dextranase (AoDex) (PDB ID [{{PDBlink}}6nzs 6NZS]).]] | ||
| − | |||
| − | |||
| + | [[File:Fig3_GH49_activesites.png|thumb|600px|right|'''Figure 3. Active sites of GH49 enzymes.''' (A-C) Molecular surfaces of Dex49A (A), IPU (B), and AoDex (C) are shown in gray. Amino acid residues and ligands are shown as stick models: catalytic residues, slate blue; residues forming active site clefts: yellow (C-terminal domain) and pink (N-terminal domain); isomaltose and isopanose, green. In (B), the isomaltose molecule (dark green) bound in N448A variant of IPU (PDB ID [{{PDBlink}}3WWG 3WWG] <cite>Miyazaki2015</cite>) is superimposed. (D) The superimposition of the catalytic residues of the three GH49 enzymes. Water molecules interacting with two general base candidates (asterisk) are shown as red sphere.]] | ||
| − | + | Three structures of GH49 enzymes are available so far <cite>Larsson2003 Mizuno2008 Ren2019</cite>, and they display a two-domain structure. The N-terminal domain is a β-sandwich and the C-terminal domain adopts a right-handed parallel β-helix. Although GH49, [[GH28]], [[GH87]], and [[GH110]] families contain enzymes with distinct substrate specificities and exhibit low overall sequence homology, they share similar β-helix folds and the three catalytic Asp residues are completely conserved <cite>Larsson2003 Mizuno2008 Itoh2020 McGuire2020</cite>. Each coil forming the cylindrical β-helix fold is composed of three β-sheets, which are named PB1, PB2, and PB3, following the original definition for a PL1 enzyme, pectate lyase C <cite>Yoder1993</cite>. | |
| − | GH49 enzymes display a two domain structure. The N-terminal domain is a β-sandwich and the C-terminal domain adopts a right-handed parallel β-helix. | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First | + | ;First gene cloning: Dextranase from ''Arthrobacter'' sp. CB-8 <cite>Okushima1991</cite>. |
| − | ;First | + | ;First stereochemistry determination: Dextranase (Dex49A) from ''Talaromyces minioluteum'' <cite>Larsson2003</cite>. |
| − | ;First general acid | + | ;First general acid residue identification: Dextranase (Dex49A) from ''Talaromyces minioluteum'' <cite>Larsson2003</cite>. |
| − | ;First 3-D structure: | + | ;First 3-D structure: Dextranase (Dex49A) from ''Talaromyces minioluteum'' by X-ray crystallography (PDB ID [{{PDBlink}}1ogm 1OGM]) <cite>Larsson2003</cite>. |
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Mizuno1999 pmid=10540747 |
| − | # | + | #Larsson2003 pmid=12962629 |
| − | # | + | #Mizuno2008 pmid=18155243 |
| − | # | + | #Akeboshi2004 pmid=15560783 |
| + | #vanSanten1999 pmid=10521427 | ||
| + | #Shimizu2002 pmid=12022868 | ||
| + | #Yoder1993 pmid=8502994 | ||
| + | #Okushima1991 pmid=1859672 | ||
| + | #Sakano1971 Sakano Y, Masuda N, and Kobayashi T. (1971). ''Hydrolysis of Pullulan by a Novel Enzyme from Aspergillus niger'', ''Agric Biol Chem'' 1971;35(6):971-973. https://doi.org/10.1271/bbb1961.35.971 | ||
| + | #Helbert2019 pmid=30850540 | ||
| + | #Miyazaki2015 pmid=25359784 | ||
| + | #Ren2019 pmid=30919632 | ||
| + | #Itoh2020 pmid=31788942 | ||
| + | #McGuire2020 pmid=33127644 | ||
| + | #Kitagawa2023 pmid=36592961 | ||
</biblio> | </biblio> | ||
[[Category:Glycoside Hydrolase Families|GH049]] | [[Category:Glycoside Hydrolase Families|GH049]] | ||
Latest revision as of 01:19, 30 September 2025
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH49 | |
| Clan | GH-N |
| Mechanism | inverting |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH49.html | |
Substrate specificities
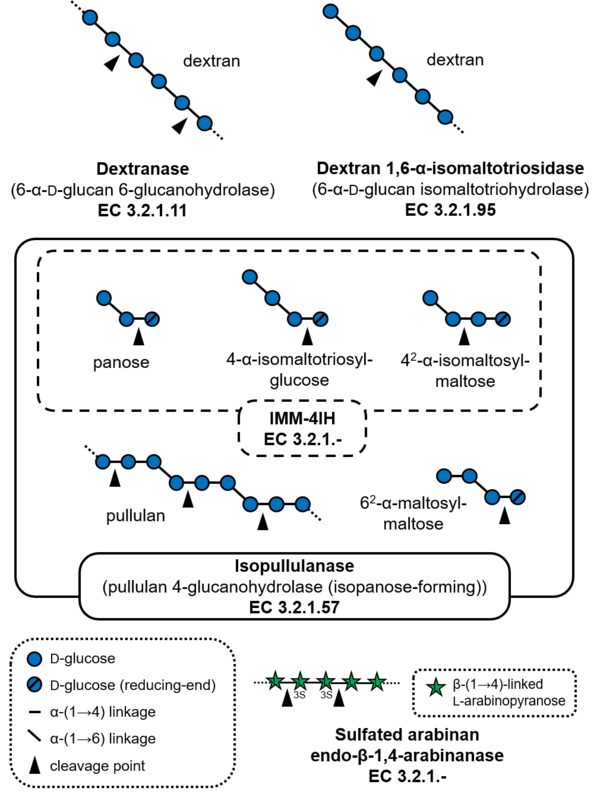
Glycoside hydrolases of GH49 cleave α-(1→6)-glucosidic linkages or α-(1→4)-glucosidic linkages of polysaccharides and oligosaccharides containing α-(1→6)-glucosidic linkages, such as dextran and pullulan. The major activities reported for this family of glycoside hydrolases are dextranase (EC 3.2.1.11), and a dextranase from Talaromyces minioluteum (formerly known as Penicillium minioluteum), Dex49A, is currently the most characterized enzyme. GH49 dextranases have been found in some bacteria and fungi. Dextran 1,6-α-isomaltotriosidase (EC 3.2.1.95) from Brevibacterium fuscum var. dextranlyticum is an exo-acting enzyme that hydrolyzes dextran from the non-reducing ends to produce isomaltotriose [1]. Isopullulanase (EC 3.2.1.57)from Aspergillus brasiliensis ATCC 9642 (formerly Aspergillus niger ATCC 9642) hydrolyzes α-(1→4)-linkages of pullulan to produce isopanose [2]. 4-O-α-D-Isomaltooligosaccharylmaltooligosaccharide 1,4-α-isomaltooligosaccharohydrolase (IMM-4IH, EC 3.2.1.-) from Sarocladium kiliense possesses more strict substrate specificity than isopullulanase and cannot hydrolyze pullulan [3]. As an exception, endo-acting sulfated-arabinan hydrolase (EC 3.2.1-) from Phocaeicola plebeius degrades sulfated arabinan produced by green algae Chaetomorpha sp. and Cladophora sp. [4].
Kinetics and Mechanism
Family GH49 α-glycosidases are inverting enzymes, as first shown by NMR on a dextranase Dex49A from Talaromyces minioluteum [5] . Optical rotation analysis of the hydrolysis of panose by isopullulanase supported the inverting mechanism [6].
Catalytic Residues
Three Asp residues (Asp376, Asp395, and Asp396 in Dex49A) are conserved in the catalytic center of clan GH-N members, GH49 and GH28 enzymes [5, 7], which is also the case for GH87 and GH110. All three of the Asp mutants of A. brasiliensis isopullulanase, lost their activities [6]. The general acid was first identified in Dex49A from Talaromyces minioluteum as Asp395 following the three-dimensional structure determination. To date, it is unclear whether either (or both) of the Asp residues (Asp376 and Asp396 in Dex49A) acts as a general base in the reaction of GH49 and GH28 enzymes [5, 8, 9].
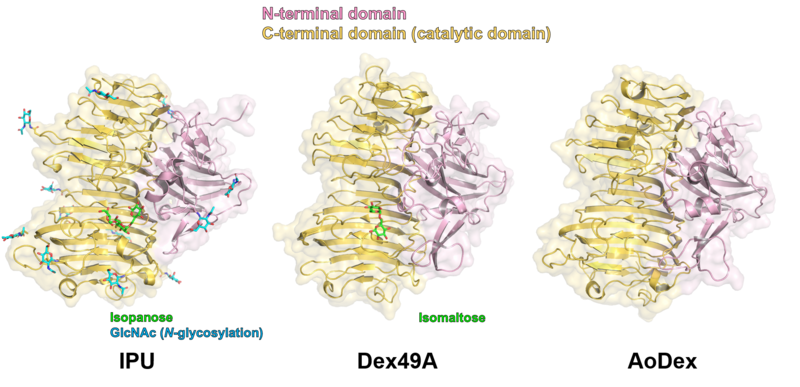
Three-dimensional structures
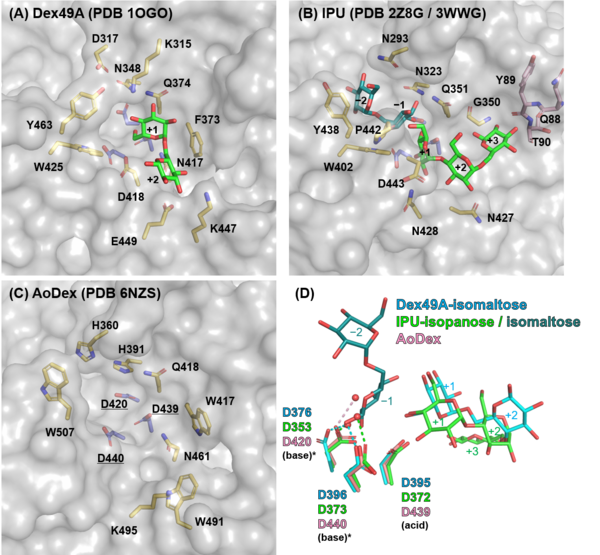
Three structures of GH49 enzymes are available so far [5, 7, 11], and they display a two-domain structure. The N-terminal domain is a β-sandwich and the C-terminal domain adopts a right-handed parallel β-helix. Although GH49, GH28, GH87, and GH110 families contain enzymes with distinct substrate specificities and exhibit low overall sequence homology, they share similar β-helix folds and the three catalytic Asp residues are completely conserved [5, 7, 12, 13]. Each coil forming the cylindrical β-helix fold is composed of three β-sheets, which are named PB1, PB2, and PB3, following the original definition for a PL1 enzyme, pectate lyase C [14].
Family Firsts
- First gene cloning
- Dextranase from Arthrobacter sp. CB-8 [15].
- First stereochemistry determination
- Dextranase (Dex49A) from Talaromyces minioluteum [5].
- First general acid residue identification
- Dextranase (Dex49A) from Talaromyces minioluteum [5].
- First 3-D structure
- Dextranase (Dex49A) from Talaromyces minioluteum by X-ray crystallography (PDB ID 1OGM) [5].
References
Error fetching PMID 30850540:
Error fetching PMID 25359784:
Error fetching PMID 30919632:
Error fetching PMID 31788942:
Error fetching PMID 33127644:
Error fetching PMID 36592961:
- Mizuno T, Mori H, Ito H, Matsui H, Kimura A, and Chiba S. (1999). Molecular cloning of isomaltotrio-dextranase gene from Brevibacterium fuscum var. dextranlyticum strain 0407 and its expression in Escherichia coli. Biosci Biotechnol Biochem. 1999;63(9):1582-8. DOI:10.1271/bbb.63.1582 |
-
Sakano Y, Masuda N, and Kobayashi T. (1971). Hydrolysis of Pullulan by a Novel Enzyme from Aspergillus niger, Agric Biol Chem 1971;35(6):971-973. https://doi.org/10.1271/bbb1961.35.971
- Error fetching PMID 36592961:
- Error fetching PMID 30850540:
- Larsson AM, Andersson R, Ståhlberg J, Kenne L, and Jones TA. (2003). Dextranase from Penicillium minioluteum: reaction course, crystal structure, and product complex. Structure. 2003;11(9):1111-21. DOI:10.1016/s0969-2126(03)00147-3 |
- Akeboshi H, Tonozuka T, Furukawa T, Ichikawa K, Aoki H, Shimonishi A, Nishikawa A, and Sakano Y. (2004). Insights into the reaction mechanism of glycosyl hydrolase family 49. Site-directed mutagenesis and substrate preference of isopullulanase. Eur J Biochem. 2004;271(22):4420-7. DOI:10.1111/j.1432-1033.2004.04378.x |
- Mizuno M, Koide A, Yamamura A, Akeboshi H, Yoshida H, Kamitori S, Sakano Y, Nishikawa A, and Tonozuka T. (2008). Crystal structure of Aspergillus niger isopullulanase, a member of glycoside hydrolase family 49. J Mol Biol. 2008;376(1):210-20. DOI:10.1016/j.jmb.2007.11.098 |
- Error fetching PMID 10521427:
- Shimizu T, Nakatsu T, Miyairi K, Okuno T, and Kato H. (2002). Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution. Biochemistry. 2002;41(21):6651-9. DOI:10.1021/bi025541a |
- Error fetching PMID 25359784:
- Error fetching PMID 30919632:
- Error fetching PMID 31788942:
- Error fetching PMID 33127644:
- Yoder MD, Keen NT, and Jurnak F. (1993). New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260(5113):1503-7. DOI:10.1126/science.8502994 |
- Okushima M, Sugino D, Kouno Y, Nakano S, Miyahara J, Toda H, Kubo S, and Matsushiro A. (1991). Molecular cloning and nucleotide sequencing of the Arthrobacter dextranase gene and its expression in Escherichia coli and Streptococcus sanguis. Jpn J Genet. 1991;66(2):173-87. DOI:10.1266/jjg.66.173 |