CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 4
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Claire Dumon^^^
- Responsible Curator: ^^^Harry Gilbert^^^
| CAZy DB link | |
| https://www.cazy.org/CBM4.html |
Ligand specificities
CBM4 is a bacterial family of protein modules that comprise around 150 amino acids. The CBM4s target primarily xylans [1], β1,3-glucans [2] or β1,4-glucans [3], although they all display a degree of non-specific binding to other glycans. For example CBM4s that primarily target xylan or β1,4-glucan also bind to β1,3-β1,4-mixed linked glucans. CBM4s that target predominantly β1,3-glucans also recognise β1,6-glucans [4]. It should also be emphasised that the CBM4s that preferentially target β1,4-glucans interact with amorphous cellulose but do not bind to crystalline cellulose [5].
CBM4s derived from thermophilic and mesophilic bacteria display affinities (KD at room temperature) for their primary ligands of ~1 and ~50 µM, respectively [2]. When measured, binding for non-primary ligands was ~100-fold lower than for the target glycan [6]. The affinity for oligosaccharides increased up to the pentaose suggesting the presence of five major sugar binding sites [1, 2, 7]. In all cases isothermal titration calorimetry showed that ligand binding was enthalpically driven and coverage of polysaccharides at saturation indicated an endo-binding mode [1, 2]. Thus CBM4 is a type B family.
Structural Features
The solution 3D structure of the C-terminal CBM4 (RmCBM4-2) of the Rhodothermus marinus xylanase Xyn10A was determined by NMR [8]. RmCBM4-2 displays a classical β-jelly-roll fold consisting of two β-sheets comprising five (β-sheet 1) and six (β-sheets) anti-parallel b strands, respectively (Figure 1). RmCBM4-2 is particularly thermostable with a Tm of 97 oC which is, in part, caused by two structural calcium ions. The location of the high affinity metal binding site is conserved in all CBMs with a β-sandwich fold [9], while the low affinity calcium site [10] has not been reported in any other family of CBMs. The ligand binding cleft is located on the concave surface (β-sheet 1) and changes in the chemical shifts of NMR spectra upon oligosaccharide titrations indicated that two aromatic residues and a number of polar amino acids interact with ligand in its 3-fold screw-axis conformation [8].
A crystal structure of a mutant of RmCBM4-2 (X-2; see the Functionalities Section below) in complex with xylopentaose was determined. The data revealed the polar and apolar interactions between the CBM and its ligand, and broadly supports predictions made previously [11]. Selection of xylan, rather than soluble b1,4-glucans reflects the requirement for a ligand with a precise 3-fold helical conformation, adopted by xylans, as opposed to the twisted orientation of the corresponding glucose polymers. Apart from one C6 hydroxymethyl group, which caused an important arginine to adopt two conformations, the substantial reduction in affinity for gluco-configured ligands was not the result of steric clashes. The structure of an engineered RmCBM4-2 with increased affinity for xyloglucan compared to the wild type protein has been subjected to extensive structural studies [11, 12, 13]. One of these studies, using neutron crystallography, revealed increased affinity for xyloglucan through the introduction of polar interactions with the xylose side chains of the ligand [12].
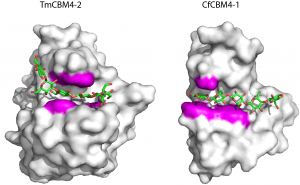
The crystal structures of TmCBM4-2 and CfCBM4-1, which bind primarily to β1,3-glucan and β1,4-glucan, respectively, were determined in complex with appropriate pentaose ligands [14]. The location of the ligand binding cleft, in common with the xylan binding RmCBM4-2, comprises β-sheet 1. A constellation of three aromatic residues, conserved in the two CBMs, form a central aromatic cradle that plays a central role in ligand recognition. These apolar contacts are augmented by a small number of hydrogen bonds with similarly conserved polar amino acids. The importance of these residues was confirmed by spectroscopic and mutagenesis data [2, 7]. Ligand specificity is conferred by the linear and U-shaped clefts of CfCBM4-1 and TmCBM4-2, respectively, that present optimal topographies for the conformations adopted by β1,4 and β1,3 glucans.
Functionalities
CBM4s are located in a variety of xylanases and endo-acting glucanases that target β1,3, mixed linked β1,3-β1,4 or β1,4 linkages. In general the specificity of CBM4s are consistent with the substrate specificity of the enzyme, although the four CBM4s in a GH16 laminarinase from Clostridium thermocellum (CtLic16A) bound to a wide range of glycans, many of which share little structural similarities [15]. An interesting example of this functional relationship between enzyme activity and CBM4 specificity is found in a GH16 laminarinase that displays activity against β1,3-glucan (3G) and mixed linked β1,3-β1,4 glucan (34G), consistent with the preference of the N-terminal CBM4 for 3G and the C-terminal CBM4 for 34G [4]. The only evidence of increased affinity of CBM4s through avidity effects is in CtLic16A [15].
Using phage display of random mutant libraries a range of specificities have been introduced into the RmCBM4-2 scaffold [16]. In addition to generating variants with increased specificity for xylan [17], mutants of the CBM4 were produced that bound specifically to xyloglucan [18, 19] and even the protein component of immunoglobulins [20]. The engineered CBM4s have been used to probe plant cell wall architectures [21, 22].
Family Firsts
- First Identified
The N-terminal CBM4 (CfCBM4-1)from the Cellulomonas fimi endoglucanase CenC [5].
- First Structural Characterization
The first structural characterization of a CBM4 was a solution NMR structure of CfCBM4-1 [23]. The first crystal structures of a CBM, and the first ligand complex of this family were CfCBM4-1 and TmCBM4-2 [14].
References
- Abou Hachem M, Nordberg Karlsson E, Bartonek-Roxâ E, Raghothama S, Simpson PJ, Gilbert HJ, Williamson MP, and Holst O. (2000). Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression and binding studies. Biochem J. 2000;345 Pt 1(Pt 1):53-60. | Google Books | Open Library
- Boraston AB, Warren RA, and Kilburn DG. (2001). beta-1,3-Glucan binding by a thermostable carbohydrate-binding module from Thermotoga maritima. Biochemistry. 2001;40(48):14679-85. DOI:10.1021/bi015760g |
- Tomme P, Creagh AL, Kilburn DG, and Haynes CA. (1996). Interaction of polysaccharides with the N-terminal cellulose-binding domain of Cellulomonas fimi CenC. 1. Binding specificity and calorimetric analysis. Biochemistry. 1996;35(44):13885-94. DOI:10.1021/bi961185i |
- Zverlov VV, Volkov IY, Velikodvorskaya GA, and Schwarz WH. (2001). The binding pattern of two carbohydrate-binding modules of laminarinase Lam16A from Thermotoga neapolitana: differences in beta-glucan binding within family CBM4. Microbiology (Reading). 2001;147(Pt 3):621-629. DOI:10.1099/00221287-147-3-621 |
- Coutinho JB, Gilkes NR, Warren RA, Kilburn DG, and Miller RC Jr. (1992). The binding of Cellulomonas fimi endoglucanase C (CenC) to cellulose and Sephadex is mediated by the N-terminal repeats. Mol Microbiol. 1992;6(9):1243-52. DOI:10.1111/j.1365-2958.1992.tb01563.x |
- Johnson PE, Tomme P, Joshi MD, and McIntosh LP. (1996). Interaction of soluble cellooligosaccharides with the N-terminal cellulose-binding domain of Cellulomonas fimi CenC 2. NMR and ultraviolet absorption spectroscopy. Biochemistry. 1996;35(44):13895-906. DOI:10.1021/bi961186a |
- Simpson PJ, Jamieson SJ, Abou-Hachem M, Karlsson EN, Gilbert HJ, Holst O, and Williamson MP. (2002). The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry. 2002;41(18):5712-9. DOI:10.1021/bi012093i |
- Boraston AB, Bolam DN, Gilbert HJ, and Davies GJ. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769-81. DOI:10.1042/BJ20040892 |
- Abou-Hachem M, Karlsson EN, Simpson PJ, Linse S, Sellers P, Williamson MP, Jamieson SJ, Gilbert HJ, Bolam DN, and Holst O. (2002). Calcium binding and thermostability of carbohydrate binding module CBM4-2 of Xyn10A from Rhodothermus marinus. Biochemistry. 2002;41(18):5720-9. DOI:10.1021/bi012094a |
- von Schantz L, Håkansson M, Logan DT, Walse B, Osterlin J, Nordberg-Karlsson E, and Ohlin M. (2012). Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules. Glycobiology. 2012;22(7):948-61. DOI:10.1093/glycob/cws063 |
- Fisher SZ, von Schantz L, Håkansson M, Logan DT, and Ohlin M. (2015). Neutron crystallographic studies reveal hydrogen bond and water-mediated interactions between a carbohydrate-binding module and its bound carbohydrate ligand. Biochemistry. 2015;54(42):6435-8. DOI:10.1021/acs.biochem.5b01058 |
- Gullfot F, Tan TC, von Schantz L, Karlsson EN, Ohlin M, Brumer H, and Divne C. (2010). The crystal structure of XG-34, an evolved xyloglucan-specific carbohydrate-binding module. Proteins. 2010;78(3):785-9. DOI:10.1002/prot.22642 |
- Boraston AB, Nurizzo D, Notenboom V, Ducros V, Rose DR, Kilburn DG, and Davies GJ. (2002). Differential oligosaccharide recognition by evolutionarily-related beta-1,4 and beta-1,3 glucan-binding modules. J Mol Biol. 2002;319(5):1143-56. DOI:10.1016/S0022-2836(02)00374-1 |
-
Dvortsov, I. A., Lunina, N. A., Zverlov, V. V., and Velikodvorskaya, G. A. (2012)Properties of four C-terminal carbohydrate-binding modules (CBM4) of laminarinase Lic16A of
Clostridium thermocellum Molecular Biology 46, 817-822 https://doi.org/10.1134/S0026893312060039
-
Gunnarsson, L. C., Karlsson, E. N., Andersson, M., Holst, O., and Ohlin, M. (2006) Molecular engineering of a thermostable carbohydrate-binding module. Biocatalysis and Biotransformation 24, 31-37 https://doi.org/10.1080/10242420500518516
- Cicortas Gunnarsson L, Montanier C, Tunnicliffe RB, Williamson MP, Gilbert HJ, Nordberg Karlsson E, and Ohlin M. (2007). Novel xylan-binding properties of an engineered family 4 carbohydrate-binding module. Biochem J. 2007;406(2):209-14. DOI:10.1042/BJ20070128 |
- von Schantz L, Gullfot F, Scheer S, Filonova L, Cicortas Gunnarsson L, Flint JE, Daniel G, Nordberg-Karlsson E, Brumer H, and Ohlin M. (2009). Affinity maturation generates greatly improved xyloglucan-specific carbohydrate binding modules. BMC Biotechnol. 2009;9:92. DOI:10.1186/1472-6750-9-92 |
- Gunnarsson LC, Zhou Q, Montanier C, Karlsson EN, Brumer H 3rd, and Ohlin M. (2006). Engineered xyloglucan specificity in a carbohydrate-binding module. Glycobiology. 2006;16(12):1171-80. DOI:10.1093/glycob/cwl038 |
- Gunnarsson LC, Dexlin L, Karlsson EN, Holst O, and Ohlin M. (2006). Evolution of a carbohydrate binding module into a protein-specific binder. Biomol Eng. 2006;23(2-3):111-7. DOI:10.1016/j.bioeng.2005.12.002 |
-
Sandquist, D., Filonova, L., von Schantz, L., Ohlin, M., and Daniel, G. (2010) Microdistribution of xyloglucan in differentiating poplar cells. Bioresources 5, 796-807
- Filonova L, Gunnarsson LC, Daniel G, and Ohlin M. (2007). Synthetic xylan-binding modules for mapping of pulp fibres and wood sections. BMC Plant Biol. 2007;7:54. DOI:10.1186/1471-2229-7-54 |
- Johnson PE, Joshi MD, Tomme P, Kilburn DG, and McIntosh LP. (1996). Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35(45):14381-94. DOI:10.1021/bi961612s |