CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Auxiliary Activity Family 5
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Maria Cleveland^^^ and ^^^Yann Mathieu^^^
- Responsible Curator: ^^^Harry Brumer^^^
| Auxiliary Activity Family AA5 | |
| Fold | Seven-bladed β-propeller |
| Mechanism | Copper Radical Oxidase |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/AA5.html | |
General Properties
Enzymes from the CAZy family AA5 are mononuclear copper-radical oxidases (CRO) that perform catalysis independently of complex organic cofactors such as FAD or NADP and use oxygen as their electron acceptor (EC 1.1.3.-).Family AA5 enzymes are classified in two subfamilies: subfamily AA5_1 contains characterized glyoxal oxidases (EC 1.2.3.15) [1] and subfamily AA5_2 contains galactose oxidases (EC 1.1.3.9) [2], as well as the more recently discovered raffinose oxidases [3, 4], aliphatic alcohol oxidases (EC 1.1.3.13) [4, 5, 6] and aryl alcohol oxidase (EC 1.1.3.7) [7, 8].
The most studied enzyme in subfamily AA5_1 is the glyoxal oxidase from Phanerochaete chrysosporium discovered in 1987 [9]. For subfamily AA5_2, the archetypal galactose-6 oxidase from Fusarium graminearum (FgrGalOx) was first reported in 1959 from cultures of Polyporus circinatus (later renamed Fusarium graminearum [10, 11]. While this first report already established FgrGalOx as a metalloenzyme; its copper requirement was later confirmed [12]. Until 2015 the characterized enzymes from the AA5_2 subfamily were found to exhibit mainly galactose oxidase activity, but since then novel non-carbohydrate oxidase enzymes were found [4, 5, 6, 7, 8].
Substrate Specificities
Content is to be added here.
Authors may get an idea of what to put in each field from Curator Approved Auxiliary Activity Families and Glycoside Hydrolase Families. (TIP: Right click with your mouse and open this link in a new browser window...)
In the meantime, please see these references for an essential introduction to the CAZy classification system: [13, 14].
Kinetics and Mechanism
Content is to be added here.
Catalytic Residues
Content is to be added here.
Three-dimensional Structures
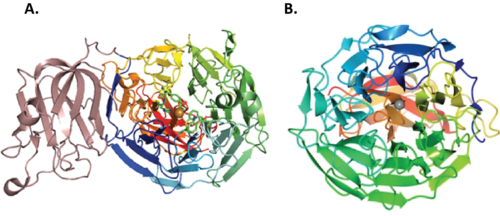
AA5 share a seven-bladed β-propeller fold [5, 7, 15] as the catalytic domain containing the active site. The archetypal FgrGalOx contains three domains: domain 1 has a “β sandwich” structure identified as a carbohydrate binding module (CBM32 REF CMB32 PAGE) with affinity for galactose, domain 2 is the catalytic domain and domain 3 is the smallest, which forms a hydrogen bonding network to stabilize domain 2 [15]. Other characterized AA5_2 enzymes from Fusarium species contain CBM32 [4, 16, 17, 18], even though some do not display canonical galactose oxidase activity (ex. FgrAAO and FoxAAO) [4, 8]. In contrast, CgrAlcOx, CglAlcOx and ChiAlcOx do not poses any CBM [5, 6], while CgrAAO and CgrRafOx have a PAN domain present instead [3, 7]. PorAlcOx contained a WSC domain that was able to bind xylans and fungal chitin/β-1,3-glucan, implicating the domains involvement in enzyme anchoring on the plant surface [6]. In addition, the fusion of a galactose oxidase with a CBM29 has shown an increase in catalytic efficiency of the construct on galactose-containing hemicelluloses compared to WT [19].
Family Firsts
- First stereochemistry determination
- Content is to be added here.
- First catalytic nucleophile identification
- Content is to be added here.
- First general acid/base residue identification
- Content is to be added here.
- First 3-D structure
- Content is to be added here.
References
- Daou M and Faulds CB. (2017). Glyoxal oxidases: their nature and properties. World J Microbiol Biotechnol. 2017;33(5):87. DOI:10.1007/s11274-017-2254-1 |
- Whittaker JW (2003). Free radical catalysis by galactose oxidase. Chem Rev. 2003;103(6):2347-63. DOI:10.1021/cr020425z |
- Andberg M, Mollerup F, Parikka K, Koutaniemi S, Boer H, Juvonen M, Master E, Tenkanen M, and Kruus K. (2017). A Novel Colletotrichum graminicola Raffinose Oxidase in the AA5 Family. Appl Environ Microbiol. 2017;83(20). DOI:10.1128/AEM.01383-17 |
-
pmid=
- Yin DT, Urresti S, Lafond M, Johnston EM, Derikvand F, Ciano L, Berrin JG, Henrissat B, Walton PH, Davies GJ, and Brumer H. (2015). Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family. Nat Commun. 2015;6:10197. DOI:10.1038/ncomms10197 |
- Oide S, Tanaka Y, Watanabe A, and Inui M. (2019). Carbohydrate-binding property of a cell wall integrity and stress response component (WSC) domain of an alcohol oxidase from the rice blast pathogen Pyricularia oryzae. Enzyme Microb Technol. 2019;125:13-20. DOI:10.1016/j.enzmictec.2019.02.009 |
-
Mathieu, Y., Offen, W. A., Forget, S. M., Ciano, L., Viborg, A. H., Blagova, E., Henrissat, B., Walton, P.H, Davies, G.J, and Brumer, H. (2020). Discovery of a fungal copper radical oxidase with high catalytic efficiency toward 5-hydroxymethylfurfural and benzyl alcohols for bioprocessing. ACS Catalysis, 10(5), 3042-3058. https://pubs.acs.org/doi/abs/10.1021/acscatal.9b04727
- Cleveland M, Lafond M, Xia FR, Chung R, Mulyk P, Hein JE, and Brumer H. (2021). Two Fusarium copper radical oxidases with high activity on aryl alcohols. Biotechnol Biofuels. 2021;14(1):138. DOI:10.1186/s13068-021-01984-0 |
- Kersten PJ and Kirk TK. (1987). Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J Bacteriol. 1987;169(5):2195-201. DOI:10.1128/jb.169.5.2195-2201.1987 |
-
Ögel, Z. B.; Brayford, D.; McPherson, M. J., (1994). Cellulose-triggered sporulation in the galactose oxidase-producing fungus Cladobotryum (Dactylium) dendroides NRRL 2903 and its re-identification as a species of Fusarium. Mycol. Res., 98 (4), 474-480. https://doi.org/10.1016/j.pep.2014.12.010
- COOPER JA, SMITH W, BACILA M, and MEDINA H. (1959). Galactose oxidase from Polyporus circinatus, Fr. J Biol Chem. 1959;234(3):445-8. | Google Books | Open Library
- AMARAL D, BERNSTEIN L, MORSE D, and HORECKER BL. (1963). Galactose oxidase of Polyporus circinatus: a copper enzyme. J Biol Chem. 1963;238:2281-4. | Google Books | Open Library
-
Davies, G.J. and Sinnott, M.L. (2008) Sorting the diverse: the sequence-based classifications of carbohydrate-active enzymes. The Biochemist, vol. 30, no. 4., pp. 26-32. Download PDF version.
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, and Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233-8. DOI:10.1093/nar/gkn663 |
- Ito N, Phillips SE, Yadav KD, and Knowles PF. (1994). Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol. 1994;238(5):794-814. DOI:10.1006/jmbi.1994.1335 |
- Paukner R, Staudigl P, Choosri W, Sygmund C, Halada P, Haltrich D, and Leitner C. (2014). Galactose oxidase from Fusarium oxysporum--expression in E. coli and P. pastoris and biochemical characterization. PLoS One. 2014;9(6):e100116. DOI:10.1371/journal.pone.0100116 |
- Paukner R, Staudigl P, Choosri W, Haltrich D, and Leitner C. (2015). Expression, purification, and characterization of galactose oxidase of Fusarium sambucinum in E. coli. Protein Expr Purif. 2015;108:73-79. DOI:10.1016/j.pep.2014.12.010 |
- Faria CB, de Castro FF, Martim DB, Abe CAL, Prates KV, de Oliveira MAS, and Barbosa-Tessmann IP. (2019). Production of Galactose Oxidase Inside the Fusarium fujikuroi Species Complex and Recombinant Expression and Characterization of the Galactose Oxidase GaoA Protein from Fusarium subglutinans. Mol Biotechnol. 2019;61(9):633-649. DOI:10.1007/s12033-019-00190-6 |
- Mollerup F and Master E. (2016). Influence of a family 29 carbohydrate binding module on the recombinant production of galactose oxidase in Pichia pastoris. Data Brief. 2016;6:176-83. DOI:10.1016/j.dib.2015.11.032 |
- Whittaker MM, Kersten PJ, Nakamura N, Sanders-Loehr J, Schweizer ES, and Whittaker JW. (1996). Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J Biol Chem. 1996;271(2):681-7. DOI:10.1074/jbc.271.2.681 |