CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Polysaccharide epimerases
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Margrethe Gaardlos^^^ and ^^^Anne Tondervik^^^
- Responsible Curator: ^^^Finn Aachmann^^^
Introduction
Classification
Mannuronan C5-epimerases
Substrate specificity
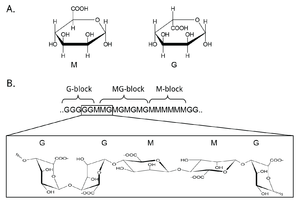
Mannuronan C5-epimerases are a group of enzymes that catalyze epimerization at the polymer-level of β-d-mannuronic acid residues (hereafter denoted M) into α-l-guluronic acid residues (hereafter denoted G) in alginate [1, 2, 3]. Alginate is an anionic polysaccharide made by brown seaweeds, some species of red algae, and the gram-negative bacterial genera Pseudomonas and Azotobacter [4, 5, 6, 7, 8]. The function of alginate in the different organisms are various, and related to structure, protection and surface adhesion [9, 10, 11, 12]. Alginate is a copolymer of the two 1-4 linked epimers [13, 14, 15], and by changing the composition of the two monomers the epimerases fine-tune the properties of the polymer [16].
At first, alginate is made as a homopolymer of M in the cell. Epimerases then convert some of the M residues in the polymer into G-residues [3, 17, 18]. This epimerization is not random and creates block structures of M, G or alternating MG [19, 20]. Alginate residues that are oxidized or acetylated are not substrates for the epimerases, and acetylation of alginate could be a way to control epimerization in nature [21, 22].
Mannuronan C5-epimerases exist both in algae and in bacteria [1, 23]. Gene analyses propose as many as 31 different genes encoding putative mannuronan C-5 epimerases in the brown algae Ectocarpus [24]. However, the algal epimerases are difficult to express and it is the bacterial enzymes that have been studied most extensively [24, 25]. Two categories of bacterial mannuronan C-5-epimerases have been described: the periplasmic AlgG and the extracellular and calcium dependent AlgE. AlgG creates single G residues in stretches of mannuronan, while the AlgE enzymes are processive and create MG-blocks and G-blocks. Pseudomonas is only known to produce AlgG [18, 26, 27], while A. vinelandii contains seven active AlgE enzymes in addition to AlgG [28, 29, 30, 31]. A mutant strain of P. fluorescens without the algG gene creates pure mannuronan [32]. This strain can be used to produce unepimerized substrate, which is useful for the study of the epimerization reaction. Methods for studying this are discussed in a later section.
Product profiles
The abundance of epimerases giving slightly different product profiles in A. vinelandii makes it possible for the bacteria to tailor alginate so it can fulfill different functions [29, 33]. This is done in three different ways. Firstly, some AlgE enzymes are only capable of creating MG-blocks, while others also create G-blocks. Secondly, different epimerases create block stretches of different lengths. Lastly, one of the known AlgE epimerases has a dual epimerase/lyase activity and thus modifies the polymer length [31]. Weak lyase activity has also been observed in other AlgEs [34, 35]. It is not certain whether this serves a function or if it is the result of failed epimerization.
Catalytic reaction
The extracellular A. vinelandii AlgE enzymes are studied extensively. They consist of different combinations of an independently catalytic module, the A-module, and a smaller R-module thought to modify binding [33, 36]. The enzymes' direction of movement along the substrate is not determined, but there are indications that they move along their polymeric substrate from the non-reducing to the reducing end [35, 37]. The epimerases show various degrees of processivity [35, 38, 39, 40], where AlgE4 catalyzes around 10-12 epimerizations before disassociating from the substrate [37, 41]. Epimerases are thought to only epimerize every other residue in one binding event, which means that the G-block formers will need to bind the MG-product of the first reaction again to form G-blocks [35, 37]. The reason why some epimerases can not form G-blocks may be related to their interactions with poly-MG [41].
Mechanism
The mechanism is suggested to be similar to the lyase mechanism, as illustrated in the figure [42]. This is supported by several of the A. vinelandii enzymes having both lyase and epimerase activity [31, 34, 35, 43]. Different versions of NNHSY is a common motif in both epimerases and lyases, and it is implicated to be important for catalysis or binding [32, 44, 45].
The proposed epimerase mechanism is initiated with neutralization of the negative charge of the carboxylate group. This is followed by abstraction of H5 by a catalytic base and ends with an addition of another proton to the opposite side of the sugar ring by a catalytic acid. The conformation of the monomer flips from 4C1 to 1C4, and changes it from β-d-mannuronate to α-l-guluronate. In the lyase mechanism, the second step is a β-elimination of the 4-O-glycosidic bond to form a 4-deoxy-l-erythro-hex-4-enepyranosyluronate, called Δ, at the non-reducing end.
Methods to study the reaction
Catalytic residues
Role of calcium
Substrate binding
Three-dimensional structures
References
- Haug A and Larsen B. (1969). Biosynthesis of alginate. Epimerisation of D-mannuronic to L-guluronic acid residues in the polymer chain. Biochim Biophys Acta. 1969;192(3):557-9. DOI:10.1016/0304-4165(69)90414-0 |
- Larsen B and Haug A. (1971). Biosynthesis of alginate. 1. Composition and structure of alginate produced by Azotobacter vinelandii (Lipman). Carbohydr Res. 1971;17(2):287-96. DOI:10.1016/s0008-6215(00)82536-7 |
- Haug A and Larsen B. (1971). Biosynthesis of alginate. II. Polymannuronic acid C-5-epimerase from Azotobacter vinelandii (Lipman). Carbohydr Res. 1971;17(2):297-308. DOI:10.1016/s0008-6215(00)82537-9 |
-
Stanford, Edw C C. (1883) On algin: a new substance obtained from some of the commoner species of marine algae. R. Anderson. NLM ID: 101217546
-
Gorin, P. A. J. and Spencer, J. F. T. (1966) Exocellular alginic acid from Azotobacter vinelandii. Canadian Journal of Chemistry vol. 44, no. 9., pp. 993-998. [1]
- Linker A and Jones RS. (1966). A new polysaccharide resembling alginic acid isolated from pseudomonads. J Biol Chem. 1966;241(16):3845-51. | Google Books | Open Library
- Govan JR, Fyfe JA, and Jarman TR. (1981). Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J Gen Microbiol. 1981;125(1):217-20. DOI:10.1099/00221287-125-1-217 |
-
Okazaki, M., K. and Furuya, K. Tsukayam and K. Nisizawa. (1982) Isolation and Identification of Alginic Acid from a Calcareous Red Alga Serraticardia maxima. Botanica Marina, vol. 25, no. 3., pp. 123-131. [1]
-
Painter, Terence J. (1983) Chapter 4 - Algal Polysaccharides. Edited by Gerald O. Aspinall. The Polysaccharides. New York: Academic Press. [1]
- Campos M, Martínez-Salazar JM, Lloret L, Moreno S, Núñez C, Espín G, and Soberón-Chávez G. (1996). Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178(7):1793-9. DOI:10.1128/jb.178.7.1793-1799.1996 |
- Pier GB, Coleman F, Grout M, Franklin M, and Ohman DE. (2001). Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69(3):1895-901. DOI:10.1128/IAI.69.3.1895-1901.2001 |
- Harmsen M, Yang L, Pamp SJ, and Tolker-Nielsen T. (2010). An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol. 2010;59(3):253-68. DOI:10.1111/j.1574-695X.2010.00690.x |
-
Hirst, E. L. and Jones, J. K. N and Jones, Winifred Osman. (1939) 389. The structure of alginic acid. Part I [in en]. Journal of the Chemical Society, The Royal Society of Chemistry. Vol. 0, pp. 1880–1885. [1]
- FISCHER FG and DORFEL H. (1955). [Polyuronic acids in brown algae]. Hoppe Seylers Z Physiol Chem. 1955;302(4-6):186-203. | Google Books | Open Library
-
Drummond, D W and Hirst, E L and Percival, Elizabeth. (1962) 232. The constitution of alginic acid. Journal of the Chemical Society, The Royal Society of Chemistry. Vol. 0, pp. 1208–1216. [1]
- Ertesvåg H, Høidal HK, Schjerven H, Svanem BI, and Valla S. (1999). Mannuronan C-5-epimerases and their application for in vitro and in vivo design of new alginates useful in biotechnology. Metab Eng. 1999;1(3):262-9. DOI:10.1006/mben.1999.0130 |
- Lin TY and Hassid WZ. (1966). Pathway of algnic acid synthesis in the marine brown alga, Fucus gardneri Silva. J Biol Chem. 1966;241(22):5284-97. | Google Books | Open Library
- Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, and Ohman DE. (1994). Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994;176(7):1821-30. DOI:10.1128/jb.176.7.1821-1830.1994 |
-
Haug, Arne and Larsen, Bjørn and Smidsrød, Olav. (1966) A study on the constitution of alginic acidby partial acid hydrolysis. Acta Chemica Scandinavica, vol. 5 (July), pp. 271–277. [1]
-
Haug, Arne and Larsen, Bjørn and Smidsrød, Olav. (1967) Studies on the Sequence of Uronic Acid Residues in Alginic Acid. Acta Chemica Scandinavica, vol. 21, pp. 691–794. [1]
-
Skjåk-Bræk, Gudmund and Larsen, Bjørn and Grasdalen, Hans. (1985) The role of O-acetyl groupsin the biosynthesis of alginate by Azotobacter vinelandii. Carbohydrate Research, vol. 145, no. 1, pp. 169–174. [1]
-
Kristiansen, Kåre A and Schirmer, Bjørn C and Aachmann, Finn L. and Skjåk-Bræk, Gudmund and Draget, Kurt I. and Christensen, Bjørn E. (2009) Novel alginates prepared by independent control of chain stiff-ness and distribution of G-residues: Structure and gelling properties. Carbohydrate Polymers, vol. 77, no.4, pp. 725–735. [1]
- Madgwick J, Haug A, and Larsen B. (1973). Polymannuronic acid 5-epimerase from the marine alga Pelvetia canaliculata (L.) Dcne. et Thur. Acta Chem Scand. 1973;27(9):3592-4. DOI:10.3891/acta.chem.scand.27-3592 |
- Fischl R, Bertelsen K, Gaillard F, Coelho S, Michel G, Klinger M, Boyen C, Czjzek M, and Hervé C. (2016). The cell-wall active mannuronan C5-epimerases in the model brown alga Ectocarpus: From gene context to recombinant protein. Glycobiology. 2016;26(9):973-983. DOI:10.1093/glycob/cww040 |
- Nyvall P, Corre E, Boisset C, Barbeyron T, Rousvoal S, Scornet D, Kloareg B, and Boyen C. (2003). Characterization of mannuronan C-5-epimerase genes from the brown alga Laminaria digitata. Plant Physiol. 2003;133(2):726-35. DOI:10.1104/pp.103.025981 |
- Chitnis CE and Ohman DE. (1990). Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J Bacteriol. 1990;172(6):2894-900. DOI:10.1128/jb.172.6.2894-2900.1990 |
- Morea A, Mathee K, Franklin MJ, Giacomini A, O'Regan M, and Ohman DE. (2001). Characterization of algG encoding C5-epimerase in the alginate biosynthetic gene cluster of Pseudomonas fluorescens. Gene. 2001;278(1-2):107-14. DOI:10.1016/s0378-1119(01)00685-0 |
- Ertesvåg H, Doseth B, Larsen B, Skjåk-Braek G, and Valla S. (1994). Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J Bacteriol. 1994;176(10):2846-53. DOI:10.1128/jb.176.10.2846-2853.1994 |
- Ertesvåg H, Høidal HK, Hals IK, Rian A, Doseth B, and Valla S. (1995). A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol Microbiol. 1995;16(4):719-31. DOI:10.1111/j.1365-2958.1995.tb02433.x |
- Rehm BH, Ertesvåg H, and Valla S. (1996). A new Azotobacter vinelandii mannuronan C-5-epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J Bacteriol. 1996;178(20):5884-9. DOI:10.1128/jb.178.20.5884-5889.1996 |
- Svanem BI, Skjåk-Braek G, Ertesvåg H, and Valla S. (1999). Cloning and expression of three new Aazotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5-epimerases. J Bacteriol. 1999;181(1):68-77. DOI:10.1128/JB.181.1.68-77.1999 |
- Gimmestad M, Sletta H, Ertesvåg H, Bakkevig K, Jain S, Suh SJ, Skjåk-Braek G, Ellingsen TE, Ohman DE, and Valla S. (2003). The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J Bacteriol. 2003;185(12):3515-23. DOI:10.1128/JB.185.12.3515-3523.2003 |
- Ertesvåg H and Valla S. (1999). The A modules of the Azotobacter vinelandii mannuronan-C-5-epimerase AlgE1 are sufficient for both epimerization and binding of Ca2+. J Bacteriol. 1999;181(10):3033-8. DOI:10.1128/JB.181.10.3033-3038.1999 |
-
Ramstad, Marit Valeur and Ellingsen, Trond E. and Josefsen, Kjell D. and Høidal, Hilde K. and Valla, Svein and Skjåk-Bræk, Gudmund and Levine, David W. (1999) Properties and action pattern of the recombinant mannuronan C-5-epimerase AlgE2. Enzyme and Microbial Technology, vol. 24, no. 10, pp. 636–646. [1]
- Holtan S, Bruheim P, and Skjåk-Braek G. (2006). Mode of action and subsite studies of the guluronan block-forming mannuronan C-5 epimerases AlgE1 and AlgE6. Biochem J. 2006;395(2):319-29. DOI:10.1042/BJ20051804 |
- Buchinger E, Knudsen DH, Behrens MA, Pedersen JS, Aarstad OA, Tøndervik A, Valla S, Skjåk-Bræk G, Wimmer R, and Aachmann FL. (2014). Structural and functional characterization of the R-modules in alginate C-5 epimerases AlgE4 and AlgE6 from Azotobacter vinelandii. J Biol Chem. 2014;289(45):31382-96. DOI:10.1074/jbc.M114.567008 |
- Campa C, Holtan S, Nilsen N, Bjerkan TM, Stokke BT, and Skjåk-Braek G. (2004). Biochemical analysis of the processive mechanism for epimerization of alginate by mannuronan C-5 epimerase AlgE4. Biochem J. 2004;381(Pt 1):155-64. DOI:10.1042/BJ20031265 |
- Høidal HK, Ertesvåg H, Skjåk-Braek G, Stokke BT, and Valla S. (1999). The recombinant Azotobacter vinelandii mannuronan C-5-epimerase AlgE4 epimerizes alginate by a nonrandom attack mechanism. J Biol Chem. 1999;274(18):12316-22. DOI:10.1074/jbc.274.18.12316 |
- Hartmann M, Holm OB, Johansen GA, Skjåk-Braek G, and Stokke BT. (2002). Mode of action of recombinant Azotobacter vinelandii mannuronan C-5 epimerases AlgE2 and AlgE4. Biopolymers. 2002;63(2):77-88. DOI:10.1002/bip.10017 |
-
Sletmoen, Marit and Skjåk-Bræk, Gudmund and Stokke, Bjørn T. (2004) Single-molecular Pair Unbinding Studies of Mannuronan C-5 Epimerase AlgE4 and Its Polymer Substrate, Biomacromolecules, American Chemical Society, vol. 5, no.4, pp. 1288–1295. DOI: 10.1021/BM0345211
- Håti AG, Aachmann FL, Stokke BT, Skjåk-Bræk G, and Sletmoen M. (2015). Energy Landscape of Alginate-Epimerase Interactions Assessed by Optical Tweezers and Atomic Force Microscopy. PLoS One. 2015;10(10):e0141237. DOI:10.1371/journal.pone.0141237 |
- Svanem BI, Strand WI, Ertesvag H, Skjåk-Braek G, Hartmann M, Barbeyron T, and Valla S. (2001). The catalytic activities of the bifunctional Azotobacter vinelandii mannuronan C-5-epimerase and alginate lyase AlgE7 probably originate from the same active site in the enzyme. J Biol Chem. 2001;276(34):31542-50. DOI:10.1074/jbc.M102562200 |
-
Yoon, Hye-Jin and Hashimoto, Wataru and Miyake, Osamu and Murata, Kousaku and Mikami, Bunzo. (2001) Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution, Journal of Molecular Biology, vol. 307, no. 1 pp. 9–16. DOI: 10.1006/jmbi.2000.4509
- Ertesvåg H, Erlien F, Skjåk-Braek G, Rehm BH, and Valla S. (1998). Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J Bacteriol. 1998;180(15):3779-84. DOI:10.1128/JB.180.15.3779-3784.1998 |
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, and Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233-8. DOI:10.1093/nar/gkn663 |
-
Davies, G.J. and Sinnott, M.L. (2008) Sorting the diverse: the sequence-based classifications of carbohydrate-active enzymes. The Biochemist, vol. 30, no. 4., pp. 26-32. Download PDF version.