CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Auxiliary Activity Family 3
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Roland Ludwig^^^ and ^^^Daniel Kracher^^^
- Responsible Curator: ^^^Roland Ludwig^^^
| Auxiliary Activity Family AA3 | |
| Clan | AA3 |
| Mechanism | FAD-dependent substrate oxidation |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/AA3.html | |
General properties and substrate specificities
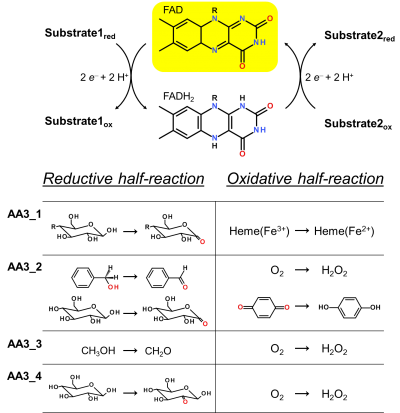
Enzymes of the CAZY family AA3 are widespread and catalyse the oxidation of alcohols or carbohydrates with the concomitant formation of hydrogen peroxide or hydroquinones [1]. AA3 enzymes are most abundant in wood-degrading fungi where they typically display a high multigenicity [2]. The main function of fungal AA3 enzymes is the stimulation of lignocellulose degradation in cooperation with other AA-enzymes such as peroxidases (AA2) [3] or lytic polysaccharide monooxygenases (AA9) [4, 5]. In yeast, AA3 enzymes are involved in the catabolism of alcohols [6], and some AA3 genes identified in insects are thought to be relevant for immunity and development [7, 8]. Recently, a bacterial AA3 enzyme with unknown biological function was isolated and characterized [9]. The functionally diverse enzymes of family AA3 all belong to the structurally related glucose-methanol-choline (GMC) family of oxidoreductases and require a flavin-adenine dinucleotide (FAD) cofactor for catalytic activity. Based on their sequences members of the AA3 family were divided into four subfamilies in the CAZy database (Figure 1). Family AA3_1 contains the flavodehydrogenase domain of cellobiose dehydrogenase (EC 1.1.99.18), family AA3_2 includes aryl alcohol oxidase (EC 1.1.3.7), glucose oxidase (EC 1.1.3.4), glucose dehydrogenase (EC 1.1.5.9) and pyranose dehydrogenase (EC 1.1.99.29), family AA3_3 consists of alcohol (methanol) oxidases (EC 1.1.3.13) and family AA3_4 comprises pyranose oxidoreductases (EC 1.1.3.10).
Subfamily AA3_1: cellobiose dehydrogenase
Cellobiose dehydrogenases (CDHs) are extracellular flavocytochromes that were first described in 1974 [10]. CDHs are exclusively found in wood-degrading and phytopathogenic fungi belonging to the phyla Basidiomycota (Class-I CDHs) and Ascomycota (Class-II and -III CDHs) [11]. They oxidize a wide variety of lignocellulose-derived saccharides to their corresponding sugar lactones. CDHs show a high preference for soluble, β-(1,4)-interlinked saccharides and scarcely oxidize monosaccharides. A common feature of all CDHs is their complex bipartite structure, which comprises a C-terminal cytochrome-binding domain (CYT) and a larger, catalytic flavodehydrogenase (DH) domain encoded within a single polypeptide chain [12] (Figure 2A). Both domains are connected by a flexible linker which typically comprises 15 – 30 amino acids. An important in vivo function of CDH is the reduction and activation of family AA9 lytic polysaccharide monooxygenases via its heme b domain [13, 14]. Recently, the holoenzyme structures of Neurospora crassa CDH (pdb: 4qi7) and Myriococcum thermophilum CDH (pdb: 4qi5) were reported. Two structures of N. crassa CDH showed an “open-state” conformation in which DH and CYT were spatially separated, whereas a structure of M. thermophilum CDH showed a “closed-state” conformation in which the propionate arm of the cytochrome domain interacted with the catalytic centre in DH [14]. Analysis by small angle scattering also suggested a number of possible intermediate conformers that exist in solution [14, 15]. While the closed-state allows interdomain electron transfer from DH to CYT, reduction of electron acceptors (e.g. AA9 enzymes) might occur in the open-state, in which the heme cofactor is fully accessible.
Subfamily AA3_2
Aryl Alcohol Oxidase/Dehydrogenase
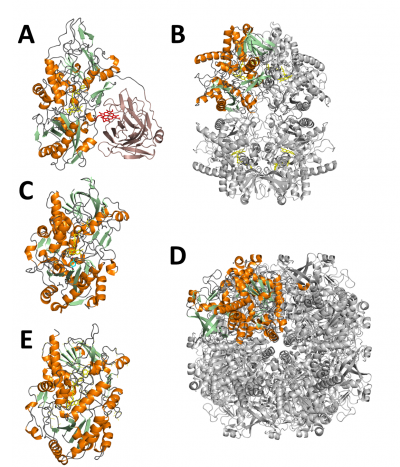
Aryl alcohol oxidoreductases (AAO) are secreted by basidiomycete fungi and proposed to play a crucial role in lignin degradation. The X-ray crystal structure of the most widely studied AAO from Pleurotus eryngii (pdb: 3fim) showed a non-covalently bound FAD cofactor in the active-site [16] (Figure 2C). Access to the active site is restricted by three aromatic residues, which interact with both the alcohol substrate and oxygen. The substrate scope of AAO includes secreted benzyl alcohols from the secondary metabolism (e.g. veratryl alcohol), or related alcohols that accumulate during lignin degradation [3]. During the reductive half-reaction, the primary alcohol of the substrate is two-electron oxidized to form the corresponding aldehyde. In addition, aromatic aldehydes can undergo further oxidation yielding the corresponding acids. The concomitantly formed hydrogen peroxide is considered essential for the activity of lignin degrading peroxidases. Recently, aryl-alcohol quinone oxidoreductases (AAQO) with a high sequence identity to Pleurotus eryngii AAO were identified in the genome of the basidiomycete Pycnoporus cinnabarinus [17]. While these enzymes showed a substrate spectrum similar to known AAOs, oxygen reduction was insignificant or very low relative to other characterised AAOs. However, reoxidation of FADH2 was very efficient with benzoquinone or phenoxy-radicals, which are formed by fungal laccases, suggesting a different functional role of AAQOs.
Glucose oxidase and glucose dehydrogenase
Glucose oxidases (GOX) (Figure 2D) and glucose dehydrogenases (GDH) catalyse the regioselective oxidation of β-D-glucose to D-glucono-1,5-lactone. Both GOX and GDH are structurally related, including a set of conserved active site residues for the binding of glucose. Therefore, a discrimination of these genes based on their sequences is difficult [2]. Glucose oxidases typically occur as homodimers whereas GDHs occur as monomers or dimers. The first crystal structure of GOX from Aspergillus niger was resolved in 1993 (pdb: 1gal [18]). A highly conserved arrangement of active-site residues, which form hydrogen bonds to all five hydroxyl groups of β-D-glucose, is responsible for the particularly high specificity towards D-glucose. The first crystal structure of GDH from Aspergillus flavus was reported in 2013, showing a slightly less conserved active site and fewer interactions with glucose (pdb: 4ynt [19]). This may also explain the promiscuous reaction of GDH with β-D-xylose, which is not observed for GOX. Both enzymes show different cosubstrate specificities. Glucose oxidases reduce atmospheric oxygen to hydrogen peroxide in their oxidative half-reaction. Proposed native functions of the enzyme are closely related to its peroxide-producing abilities and include the preservation of honey, microbial defence as well as the support of H2O2-dependent ligninases in wood degrading fungi [20]. GDHs interact very slowly with oxygen and prefer electron acceptors like quinones or phenoxy-radicals. The detailed physiological role of GDH remains elusive, but has been previously linked to the neutralization of plant laccase activity by fungi during plant infection [21].
References
- Levasseur A, Drula E, Lombard V, Coutinho PM, and Henrissat B. (2013). Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6(1):41. DOI:10.1186/1754-6834-6-41 |
- Sützl L, Laurent CVFP, Abrera AT, Schütz G, Ludwig R, and Haltrich D. (2018). Multiplicity of enzymatic functions in the CAZy AA3 family. Appl Microbiol Biotechnol. 2018;102(6):2477-2492. DOI:10.1007/s00253-018-8784-0 |
- Hernández-Ortega A, Ferreira P, and Martínez AT. (2012). Fungal aryl-alcohol oxidase: a peroxide-producing flavoenzyme involved in lignin degradation. Appl Microbiol Biotechnol. 2012;93(4):1395-410. DOI:10.1007/s00253-011-3836-8 |
- Kracher D, Scheiblbrandner S, Felice AK, Breslmayr E, Preims M, Ludwicka K, Haltrich D, Eijsink VG, and Ludwig R. (2016). Extracellular electron transfer systems fuel cellulose oxidative degradation. Science. 2016;352(6289):1098-101. DOI:10.1126/science.aaf3165 |
- Garajova S, Mathieu Y, Beccia MR, Bennati-Granier C, Biaso F, Fanuel M, Ropartz D, Guigliarelli B, Record E, Rogniaux H, Henrissat B, and Berrin JG. (2016). Single-domain flavoenzymes trigger lytic polysaccharide monooxygenases for oxidative degradation of cellulose. Sci Rep. 2016;6:28276. DOI:10.1038/srep28276 |
- Goswami P, Chinnadayyala SS, Chakraborty M, Kumar AK, and Kakoti A. (2013). An overview on alcohol oxidases and their potential applications. Appl Microbiol Biotechnol. 2013;97(10):4259-75. DOI:10.1007/s00253-013-4842-9 |
- Iida K, Cox-Foster DL, Yang X, Ko WY, and Cavener DR. (2007). Expansion and evolution of insect GMC oxidoreductases. BMC Evol Biol. 2007;7:75. DOI:10.1186/1471-2148-7-75 |
- Sun W, Shen YH, Yang WJ, Cao YF, Xiang ZH, and Zhang Z. (2012). Expansion of the silkworm GMC oxidoreductase genes is associated with immunity. Insect Biochem Mol Biol. 2012;42(12):935-45. DOI:10.1016/j.ibmb.2012.09.006 |
-
Mendes S, Banha C, Madeira J, Santos D, Miranda V, Manzanera M, Ventura MR, van Berkel WJH, Martins LO. Characterization of a bacterial pyranose 2-oxidase from Arthrobacter siccitolerans. 2016 J Mol Catal B Enzym 133, S34–S43
-
Westermark U, Eriksson KE. Cellobiose:Quinone Oxidoreductase, a New Wood-degrading Enzyme from White-rot Fungi. 1974 Acta Chem Scand 1974 28b:209–214.
-
Kracher D, Ludwig R. Cellobiose dehydrogenase: An essential enzyme for lignocellulose degradation in nature - A review. 2016 Bodenkultur 67:145–163.
- Hallberg BM, Henriksson G, Pettersson G, Vasella A, and Divne C. (2003). Mechanism of the reductive half-reaction in cellobiose dehydrogenase. J Biol Chem. 2003;278(9):7160-6. DOI:10.1074/jbc.M210961200 |
- Phillips CM, Beeson WT, Cate JH, and Marletta MA. (2011). Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6(12):1399-406. DOI:10.1021/cb200351y |
- Tan TC, Kracher D, Gandini R, Sygmund C, Kittl R, Haltrich D, Hällberg BM, Ludwig R, and Divne C. (2015). Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat Commun. 2015;6:7542. DOI:10.1038/ncomms8542 |
- Bodenheimer AM, O'Dell WB, Oliver RC, Qian S, Stanley CB, and Meilleur F. (2018). Structural investigation of cellobiose dehydrogenase IIA: Insights from small angle scattering into intra- and intermolecular electron transfer mechanisms. Biochim Biophys Acta Gen Subj. 2018;1862(4):1031-1039. DOI:10.1016/j.bbagen.2018.01.016 |