CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Polysaccharide Lyase Family 22"
Wade Abbott (talk | contribs) |
Wade Abbott (talk | contribs) |
||
| Line 32: | Line 32: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | Family 22 Polysaccharide Lyases (PL22s) contain two subfamilies <cite>Lombard2010</cite> | + | Family 22 Polysaccharide Lyases (PL22s) contain two subfamilies and several outlier sequences <cite>Lombard2010</cite>. Originally referred to as oligogalacturonide trans-eliminases (OGTE)<cite>Moran1968</cite>, PL22s are now commonly referred to as oligogalacturonide lyases (OGLs). |
As the name suggests, OGLs are preferentially active on short chain oligomers of galacturonides. PL22s are preferentially active on digalacturonate and Δ4,5-unsaturated digalacturonate <cite>Abbott2010</cite><cite>Kester1999</cite>. Activity on trigalacturonate has been shown to be significantly lower than on digalacturonate and although activity on the unsaturated dimer was lower than that of the saturated dimer, activity on Δ4,5-unsaturated trigalacturonate is comparable or higher than that of saturated trigalacturonate <cite>Kester1999</cite>. OGLs lack activity on long chain polymers of α-(1,4)-linked polygalacturonate. Activity has been demonstrated on methylated short chain galacturonides with differing levels of activity depending on the location of methylation <cite>Kester1999</cite>. Activity on 1-methyl digalacturonate was only half of what was seen on digalacturonate and no activity was found on 2-methyl digalacturonate. A similar trend was shown on trigalacturonate as well with roughly half the activity on 1-methyl trigalacturonate as on trigalacturonate and no activity on 2-methyl galacturonate. | As the name suggests, OGLs are preferentially active on short chain oligomers of galacturonides. PL22s are preferentially active on digalacturonate and Δ4,5-unsaturated digalacturonate <cite>Abbott2010</cite><cite>Kester1999</cite>. Activity on trigalacturonate has been shown to be significantly lower than on digalacturonate and although activity on the unsaturated dimer was lower than that of the saturated dimer, activity on Δ4,5-unsaturated trigalacturonate is comparable or higher than that of saturated trigalacturonate <cite>Kester1999</cite>. OGLs lack activity on long chain polymers of α-(1,4)-linked polygalacturonate. Activity has been demonstrated on methylated short chain galacturonides with differing levels of activity depending on the location of methylation <cite>Kester1999</cite>. Activity on 1-methyl digalacturonate was only half of what was seen on digalacturonate and no activity was found on 2-methyl digalacturonate. A similar trend was shown on trigalacturonate as well with roughly half the activity on 1-methyl trigalacturonate as on trigalacturonate and no activity on 2-methyl galacturonate. | ||
Revision as of 05:46, 9 September 2014
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Richard McLean^^^ and ^^^Wade Abbott^^^
- Responsible Curator: ^^^Wade Abbott^^^
| Polysaccharide Lyase Family PL22 | |
| 3D Structure | β7 propeller |
| Mechanism | β-elimination |
| Charge neutraliser | manganese |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/PL22.html | |
Substrate specificities
Family 22 Polysaccharide Lyases (PL22s) contain two subfamilies and several outlier sequences [1]. Originally referred to as oligogalacturonide trans-eliminases (OGTE)[2], PL22s are now commonly referred to as oligogalacturonide lyases (OGLs).
As the name suggests, OGLs are preferentially active on short chain oligomers of galacturonides. PL22s are preferentially active on digalacturonate and Δ4,5-unsaturated digalacturonate [3][4]. Activity on trigalacturonate has been shown to be significantly lower than on digalacturonate and although activity on the unsaturated dimer was lower than that of the saturated dimer, activity on Δ4,5-unsaturated trigalacturonate is comparable or higher than that of saturated trigalacturonate [4]. OGLs lack activity on long chain polymers of α-(1,4)-linked polygalacturonate. Activity has been demonstrated on methylated short chain galacturonides with differing levels of activity depending on the location of methylation [4]. Activity on 1-methyl digalacturonate was only half of what was seen on digalacturonate and no activity was found on 2-methyl digalacturonate. A similar trend was shown on trigalacturonate as well with roughly half the activity on 1-methyl trigalacturonate as on trigalacturonate and no activity on 2-methyl galacturonate.
Kinetics and Mechanism
Catalytic Residues
The Brønstead base for PL22s is predicted to be a histidine [3]. H242 in YE1876 from Yersinia enterocolitica subsp. enterocolitica 8081 was the first and is to date, the only catalytic residue determined reported to be in proximity to the α-proton of galacturonate. This histidine is highly conserved within Family 22 lyases with only Candidatus Solibacter usitatus Ellin6076 (gi|116225114|) displaying a threonine (T236); however, whether this gene product functions as a lyase has yet to be determined.
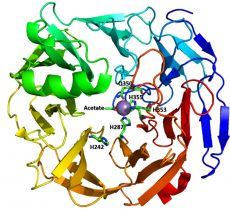
The metal coordination pocket houses a manganese ion and is comprised of three histidines (VPA0088 H287, H353, H355; YeOGL H287, H353, H355) and one glutamine (VPA0088 Q350; YeOGL Q350). It is of note however, that although these residues are perfectly conserved in all reported subfamily 1 sequences and several outlier sequences, subfamily 2 or archaeal sequences display different signatures [1]. The archaeal sequences have conserved histidines but there is variations Q350. In subfamily 2, H287 is invariant; however, Q350 is not conserved and H353 and H355 have been replaced with a glutamate and asparagine respectively. These modifications likely alter the chemistry of metal coordination selectivity.
Three-dimensional structures
The first structure of a PL22 determined was the Vibrio parahaemolyticus RIMD 2210633 (PDB 3C5M) solved in 2008 by x-ray diffraction to 2.60 Å (http://www.nesg.org/, Northeast Structural Genomics Consortium). This was followed in 2010 by Yersinia enterocolitica subsp. enterocolitica 8081 (PDB 3PE7) which was solved in complex with Mn2+ and acetate by x-ray diffraction to 1.65 Å. The two proteins share ~69% sequence identity and highly similar 3D structures.
Family Firsts
- First catalytic activity
- OGTE from Pectobacterium carotovorum ICPB EC153 (previously Erwinia carotovora). [2]
- First catalytic base identification
- YeOGL (YE1876) H242 from Yersinia enterocolitica subsp. enterocolitica 8081. [3]
- First catalytic divalent cation identification
- OGL (Dda3937_03686) from Dickeya Dadantii 3937 (previously Erwinia chrysanthemi 3937). [5].
- First 3-D structure
- VPA0088 from Vibrio parahaemolyticus RIMD 2210633. (PDB 3C5M)
References
Error fetching PMID 16593039:
Error fetching PMID 10601263:
Error fetching PMID 5671040:
Error fetching PMID 3623103:
Error fetching PMID 2695393:
Error fetching PMID 10383957:
Error fetching PMID 17189441:
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, and Henrissat B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432(3):437-44. DOI:10.1042/BJ20101185 |
- Error fetching PMID 5671040:
- Error fetching PMID 20851883:
- Error fetching PMID 10601263:
- Error fetching PMID 10383957:
- Error fetching PMID 16593039:
- Error fetching PMID 3623103:
- Error fetching PMID 2695393:
- Error fetching PMID 17189441: