CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 112"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (17 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {{CuratorApproved}} | ||
* [[Author]]: [[User:Motomitsu Kitaoka|Motomitsu Kitaoka]] | * [[Author]]: [[User:Motomitsu Kitaoka|Motomitsu Kitaoka]] | ||
| − | * [[Responsible Curator]]: [[User: | + | * [[Responsible Curator]]: [[User:Shinya Fushinobu|Shinya Fushinobu]] |
---- | ---- | ||
| Line 19: | Line 20: | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
|- | |- | ||
| − | | colspan="2" | | + | | colspan="2" |{{CAZyDBlink}}GH112.html |
|} | |} | ||
</div> | </div> | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | This family contains beta-galactoside | + | This family of [[glycoside hydrolases]] contains [[phosphorylases]]; beta-galactoside phosphorylase, β-1,3-D-galactosyl-D-hexososamine phosphorylase (EC [{{EClink}}2.4.1.211 2.4.1.211]) <cite>DerensyDron1999</cite> and β-1,4-D-galactosyl-L-rhamnose phosphorylase (EC 2.4.1.-) <cite>Nakajima2009</cite>. The former enzymes are subcategorized into galacto-''N''-biose phosphorylase, (GNBP) <cite>NakajimaAMB2008</cite>, lacto-''N''-biose I phosphorylase (LNBP) <cite>NakajimaAEM2008</cite>, and galacto-''N''-biose/lacto-''N''-biose I phosphorylase (GLNBP) <cite>DerensyDron1999 NakajimaAEM2008 Kitaoka2005</cite> based on the substrate preference on galacto-''N''-biose (GNB, Gal-β1,3-GalNAc) and lacto-''N''-biose I (LNB, Gal-β1,3-GlcNAc) <cite>NakajimaAEM2008</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | Phosphorolysis by GH112 enzymes proceeds with inversion of anomeric configuration, as first shown by Derensy-Dron et al. <cite> | + | Phosphorolysis by GH112 enzymes proceeds with inversion of anomeric configuration, as first shown by Derensy-Dron et al. <cite>DerensyDron1999</cite> on β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium bifidum'', i.e. LNB + Pi ↔ α-galactose 1-phosphate + GlcNAc. Considering the topology of the active site structure, the reaction mechanism for [[inverting]] phosphorylase is proposed to be similar to that for [[inverting]] glycoside hydrolases. With the aid of general acid residue, the enzymatic phosphorolysis occurs through direct nucleophilic attack by phosphate on the anomeric C-1 carbon, instead of the water molecule activated by a general base residue in the [[Glycoside Hydrolases#Inverting glycoside hydrolases | inverting glycoside hydrolase]] reaction. |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | The | + | The [[general acid]] residue was firstly estimated by mutational analysis on β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium longum'' <cite>Nishimoto2007</cite>. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
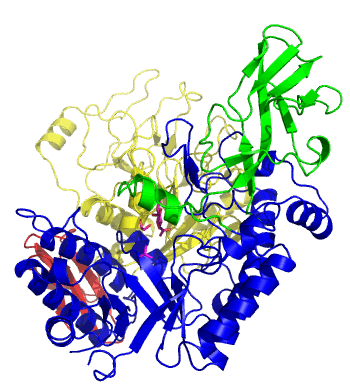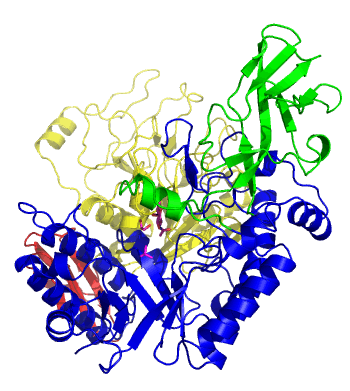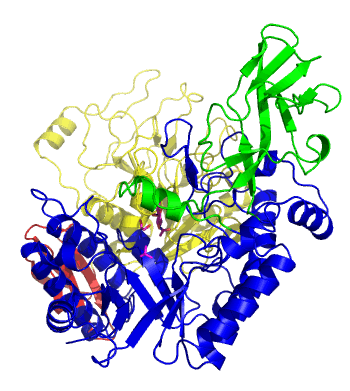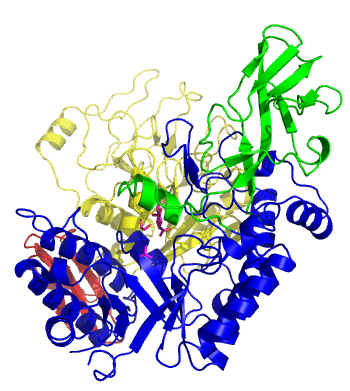
| − | The first solved 3-D structure was β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium longum'' <cite> | + | [[Image:GLNBPmov.gif|'''Figure 1:''' Movement of the catalytic TIM barrel domain induced by substrate binding. <span style="color:#0000ff"> The TIM barrel domain is in blue</span>, <span style="color:#00cc00"> an IG-like domain is in green </span>, <span style="color:#999900"> an α/β domain is in yellow </span> and <span style="color:#ff0000"> a C-terminal domain is in red </span>.|frame|right]] |
| + | The first solved 3-D structure was β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium longum'' (PDB [{{PDBlink}}2zuv 2ZUV] in complex with GlcNAc, ethylene glycol, and nitrate) <cite>Hidaka2009</cite>. The catalytic domain consists of a partially broken TIM barrel fold that is structurally similar to a β-galactosidase of [[Glycoside Hydrolase Family 42]], supporting the classification of these phosphorolytic enzymes as one of the GH families. A large conformational change in TIM barrel scaffold was observerd with substrate-binding ('''Figure 1'''). | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First sterochemistry determination: β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium bifidum'' <cite> | + | ;First sterochemistry determination: β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium bifidum'' <cite>DerensyDron1999</cite>. |
| − | ;First | + | ;First sequence identification: β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium longum'' <cite>Kitaoka2005</cite>. |
| − | ;First | + | ;First [[general acid]] residue identification: β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium longum'' <cite>Nishimoto2007</cite>. |
| − | + | ;First 3-D structure: β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium longum'' <cite>Hidaka2009</cite>. | |
| − | ;First 3-D structure: β-1,3-D-galactosyl-D-hexososamine phosphorylase from ''Bifidobacterium longum'' <cite> | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #DerensyDron1999 pmid=9889079 |
| − | # | + | #Nakajima2009 pmid=19491100 |
| − | # | + | #NakajimaAMB2008 pmid=18183385 |
| − | # | + | #NakajimaAEM2008 pmid=18723650 |
| − | # | + | #Kitaoka2005 pmid=15933016 |
| − | # | + | #Nishimoto2007 pmid=17587697 |
| − | # | + | #Hidaka2009 pmid=19124470 |
| − | |||
</biblio> | </biblio> | ||
[[Category:Glycoside Hydrolase Families|GH112]] | [[Category:Glycoside Hydrolase Families|GH112]] | ||
Latest revision as of 13:15, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family 112 | |
| Clan | none |
| Mechanism | inverting |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH112.html | |
Substrate specificities
This family of glycoside hydrolases contains phosphorylases; beta-galactoside phosphorylase, β-1,3-D-galactosyl-D-hexososamine phosphorylase (EC 2.4.1.211) [1] and β-1,4-D-galactosyl-L-rhamnose phosphorylase (EC 2.4.1.-) [2]. The former enzymes are subcategorized into galacto-N-biose phosphorylase, (GNBP) [3], lacto-N-biose I phosphorylase (LNBP) [4], and galacto-N-biose/lacto-N-biose I phosphorylase (GLNBP) [1, 4, 5] based on the substrate preference on galacto-N-biose (GNB, Gal-β1,3-GalNAc) and lacto-N-biose I (LNB, Gal-β1,3-GlcNAc) [4].
Kinetics and Mechanism
Phosphorolysis by GH112 enzymes proceeds with inversion of anomeric configuration, as first shown by Derensy-Dron et al. [1] on β-1,3-D-galactosyl-D-hexososamine phosphorylase from Bifidobacterium bifidum, i.e. LNB + Pi ↔ α-galactose 1-phosphate + GlcNAc. Considering the topology of the active site structure, the reaction mechanism for inverting phosphorylase is proposed to be similar to that for inverting glycoside hydrolases. With the aid of general acid residue, the enzymatic phosphorolysis occurs through direct nucleophilic attack by phosphate on the anomeric C-1 carbon, instead of the water molecule activated by a general base residue in the inverting glycoside hydrolase reaction.
Catalytic Residues
The general acid residue was firstly estimated by mutational analysis on β-1,3-D-galactosyl-D-hexososamine phosphorylase from Bifidobacterium longum [6].
Three-dimensional structures
The first solved 3-D structure was β-1,3-D-galactosyl-D-hexososamine phosphorylase from Bifidobacterium longum (PDB 2ZUV in complex with GlcNAc, ethylene glycol, and nitrate) [7]. The catalytic domain consists of a partially broken TIM barrel fold that is structurally similar to a β-galactosidase of Glycoside Hydrolase Family 42, supporting the classification of these phosphorolytic enzymes as one of the GH families. A large conformational change in TIM barrel scaffold was observerd with substrate-binding (Figure 1).
Family Firsts
- First sterochemistry determination
- β-1,3-D-galactosyl-D-hexososamine phosphorylase from Bifidobacterium bifidum [1].
- First sequence identification
- β-1,3-D-galactosyl-D-hexososamine phosphorylase from Bifidobacterium longum [5].
- First general acid residue identification
- β-1,3-D-galactosyl-D-hexososamine phosphorylase from Bifidobacterium longum [6].
- First 3-D structure
- β-1,3-D-galactosyl-D-hexososamine phosphorylase from Bifidobacterium longum [7].
References
- Derensy-Dron D, Krzewinski F, Brassart C, and Bouquelet S. (1999). Beta-1,3-galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol Appl Biochem. 1999;29 ( Pt 1):3-10. | Google Books | Open Library
- Nakajima M, Nishimoto M, and Kitaoka M. (2009). Characterization of three beta-galactoside phosphorylases from Clostridium phytofermentans: discovery of d-galactosyl-beta1->4-l-rhamnose phosphorylase. J Biol Chem. 2009;284(29):19220-7. DOI:10.1074/jbc.M109.007666 |
- Nakajima M, Nihira T, Nishimoto M, and Kitaoka M. (2008). Identification of galacto-N-biose phosphorylase from Clostridium perfringens ATCC13124. Appl Microbiol Biotechnol. 2008;78(3):465-71. DOI:10.1007/s00253-007-1319-8 |
- Nakajima M and Kitaoka M. (2008). Identification of lacto-N-Biose I phosphorylase from Vibrio vulnificus CMCP6. Appl Environ Microbiol. 2008;74(20):6333-7. DOI:10.1128/AEM.02846-07 |
- Kitaoka M, Tian J, and Nishimoto M. (2005). Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71(6):3158-62. DOI:10.1128/AEM.71.6.3158-3162.2005 |
- Nishimoto M and Kitaoka M. (2007). Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211). Biosci Biotechnol Biochem. 2007;71(6):1587-91. DOI:10.1271/bbb.70064 |
- Hidaka M, Nishimoto M, Kitaoka M, Wakagi T, Shoun H, and Fushinobu S. (2009). The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: a large deformation of a TIM barrel scaffold. J Biol Chem. 2009;284(11):7273-83. DOI:10.1074/jbc.M808525200 |