CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 2"
| Line 19: | Line 19: | ||
The vast majority of family 2 CBM (CBM2) members, ~95%, are bacterial in origin. These protein modules have been shown to bind to crystalline cellulose <cite>Gilkes1988</cite>, insoluble chitin <cite>Nakamura2008</cite> or xylan <cite>Black1995</cite>. The xylan-specific CBM2 modules bind only to the hemicellulose, while some cellulose-binding modules also recognize chitin. The CBM2 modules from some chitinases show a strong preference for crystalline chitin <cite>Nakamura2008</cite>. Cellulose-specific CBM2 members were also shown to bind to xyloglucan, albeit with affinities ~1000x lower than for crystalline cellulose <cite>Hernandez-Gomez2015 </cite>. Extremely weak binding to cellulooligosaccharides was detected in cellulose binding CBM2s using NMR <cite>Bolam1998</cite>. In contrast affinities of xylan binding CBM2s for their polysaccharide or oligosaccharide ligands with a degree of polymerization of six were similar <cite>Bolam2001</cite>. | The vast majority of family 2 CBM (CBM2) members, ~95%, are bacterial in origin. These protein modules have been shown to bind to crystalline cellulose <cite>Gilkes1988</cite>, insoluble chitin <cite>Nakamura2008</cite> or xylan <cite>Black1995</cite>. The xylan-specific CBM2 modules bind only to the hemicellulose, while some cellulose-binding modules also recognize chitin. The CBM2 modules from some chitinases show a strong preference for crystalline chitin <cite>Nakamura2008</cite>. Cellulose-specific CBM2 members were also shown to bind to xyloglucan, albeit with affinities ~1000x lower than for crystalline cellulose <cite>Hernandez-Gomez2015 </cite>. Extremely weak binding to cellulooligosaccharides was detected in cellulose binding CBM2s using NMR <cite>Bolam1998</cite>. In contrast affinities of xylan binding CBM2s for their polysaccharide or oligosaccharide ligands with a degree of polymerization of six were similar <cite>Bolam2001</cite>. | ||
| − | Some enzymes contain multiple CBM2 modules that, with respect to xylan, show enhanced affinity through avidity effects <cite>Bolam1998</cite>. Isothermal titration calorimetry showed that CBM2 modules that interact with their target cellulose or xylan ligands were mediated by entropic <cite>Creagh1996</cite> and enthalpic <cite>Bolam1998</cite> forces, respectively. These differences in thermodynamic forces driving ligand recognition was proposed to reflect the conformational freedom of cellulose and xylan in free solution and when bound to CBMs. Family 2 is thus highly unusual, containing both cellulose binding type A and xylan binding [[Carbohydrate-binding_modules#Types|type B]] CBMs. Labelling of type A crystalline cellulose specific CBMs, including those in CBM2, which all present three aromatic planar surface residues, showed differences in specificity for pure forms of crystalline cellulose and plant cell walls <cite>Blake2006,McLean2002</cite>. | + | Some enzymes contain multiple CBM2 modules that, with respect to xylan, show enhanced affinity through avidity effects <cite>Bolam1998</cite>. Isothermal titration calorimetry showed that CBM2 modules that interact with their target cellulose or xylan ligands were mediated by entropic <cite>Creagh1996</cite> and enthalpic <cite>Bolam1998</cite> forces, respectively. These differences in thermodynamic forces driving ligand recognition was proposed to reflect the conformational freedom of cellulose and xylan in free solution and when bound to CBMs. Family 2 is thus highly unusual, containing both cellulose binding [[Carbohydrate-binding_modules#Types|type A]] and xylan binding [[Carbohydrate-binding_modules#Types|type B]] CBMs. Labelling of type A crystalline cellulose specific CBMs, including those in CBM2, which all present three aromatic planar surface residues, showed differences in specificity for pure forms of crystalline cellulose and plant cell walls <cite>Blake2006,McLean2002</cite>. |
== Structural Features == | == Structural Features == | ||
Revision as of 07:58, 6 March 2018
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Harry Gilbert^^^
- Responsible Curator: ^^^Elizabeth Ficko-Blean^^^
| CAZy DB link | |
| https://www.cazy.org/CBM2.html |
Ligand specificities
The vast majority of family 2 CBM (CBM2) members, ~95%, are bacterial in origin. These protein modules have been shown to bind to crystalline cellulose [1], insoluble chitin [2] or xylan [3]. The xylan-specific CBM2 modules bind only to the hemicellulose, while some cellulose-binding modules also recognize chitin. The CBM2 modules from some chitinases show a strong preference for crystalline chitin [2]. Cellulose-specific CBM2 members were also shown to bind to xyloglucan, albeit with affinities ~1000x lower than for crystalline cellulose [4]. Extremely weak binding to cellulooligosaccharides was detected in cellulose binding CBM2s using NMR [5]. In contrast affinities of xylan binding CBM2s for their polysaccharide or oligosaccharide ligands with a degree of polymerization of six were similar [6].
Some enzymes contain multiple CBM2 modules that, with respect to xylan, show enhanced affinity through avidity effects [5]. Isothermal titration calorimetry showed that CBM2 modules that interact with their target cellulose or xylan ligands were mediated by entropic [7] and enthalpic [5] forces, respectively. These differences in thermodynamic forces driving ligand recognition was proposed to reflect the conformational freedom of cellulose and xylan in free solution and when bound to CBMs. Family 2 is thus highly unusual, containing both cellulose binding type A and xylan binding type B CBMs. Labelling of type A crystalline cellulose specific CBMs, including those in CBM2, which all present three aromatic planar surface residues, showed differences in specificity for pure forms of crystalline cellulose and plant cell walls [8, 9].
Structural Features
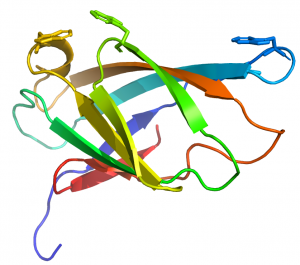
CBM2 modules generally comprise 90 to 100 residues that adopt a canonical β-jelly-roll fold (see PDB ID 1exg for an example) [10]. The two β-sheets of these CBMs consist of four (face 1) and five (face 2) antiparallel β-strands, respectively. The structure is sometimes stabilized by a disulphide bridge between the N- and C-terminal regions, but not in all cases. In cellulose binding CBM2 members, face 1 presents a planar surface that contains three highly conserved aromatic residues (generally tryptophans although tyrosine can also be present) that are positioned to bind to glucose units in linear planar cellulose chains present in the crystalline polysaccharide (Figure 1). The corresponding aromatic residues interact with N-acetylglucosamine residues in chitin [2]. Mutagenesis and chemical modfication studies confirmed the critical role played by the aromatic residues in cellulose recognition [11, 12, 13]. The central tryptophan plays a more important role in ligand binding than the distal aromatic amino acids [12].
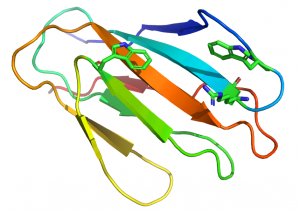
In xylan binding CBM2 modules (see PDB ID 2xbd for an example) face 1 displays a slightly concave surface that contains only two tryptophan residues [14]. The perpendicular orientation (with respect to each other; Figure 2) and location of these two tryptophan residues indicate that they interact with xylose residues n and n+2 in the three fold-helical structure of xylan chains. NMR titration experiments with xylooligosaccharides and mutagenesis studies showed that both aromatic residues play a critical role in xylan recognition.
An arginine residue adjacent to one of the tryptophans in the xylan binding CBM2 forces it into the perpendicular orientation with respect to the other surface aromatic residue (Figure 2). In cellulose binding CBM2s the corresponding perpendicular tryptophan (in xylan binding CBM2s) is planar with the other two surface aromatic residues (see PDB ID 1e5b). The adoption of the planar orientation was predicted to be mediated by the replacement of the arginine (in xylan binding CBM2s) with glycine in cellulose specific CBM2s. This was supported by mutagenesis studies showing that substituting the arginine with glycine in a xylan binding CBM2 created a planar orientation for both surface tryptophans (see Fig. 5 and 6 in [15]), abrogated xylan binding and introduced cellulose recognition [15]. Mutagenesis studies showed that alanine substitution of polar residues predicted to interact with CBM2 ligands did not influence affinity for xylan or cellulose, demonstrating that hydrophobic forces dominated carbohydrate recognition [12, 16].
Functionalities
CBM2 modules that target cellulose as the principle ligand have been reported in cellulases [1], xylan degrading enzymes (xylanases, arabinofuranosidases, esterases) [17, 18], pectate lyases [19], mannanases [20] and lytic polysaccharide monooxygenases (LPMOs) [21, 22]. Xylan and chitin binding CBM2s are restricted to enzymes that target these polysaccharides [2, 3]. Many of these modular enzymes contain other CBM families in addition to CBM2 members [23], with CBM10-CBM2 modular motifs being particularly common in glycoside hydrolases from Cellvibrio japonicus [24] (e.g. [25]) and Saccharophagus degradans [26]. CBM2 modules are linked to catalytic modules by linker sequences that are required to be >15 residues in length to maximize the boost in activity provided by the CBM [27, 28].
CBM2 modules substantially potentiate the activity of cellulases and chitinsaes against purified insoluble or crystalline forms of cellulose and chitin, respectively [1, 2]. It has been proposed that cellulase-derived CBM2 modules enhance enzyme activity by increasing the local concentration or proximity of the enzyme in the vicinity of the substrate [5]. It has, however, also been reported that a CBM2 module was able to disrupt the structure of crystalline cellulose leading to enhanced enzyme activity [29]. Cell wall imaging studies have also shown that cellulose and xylan binding CBM2s enhance the activity of a pectate lyase, arabinofuranosidase and xylanases against plant cell walls [30]. Although CBM2 modules can bind irreversibly to crystalline cellulose [31], fluorescent bleaching experiments showed that these protein modules can migrate across the surface of the polysaccharide [32]. CBM2 protein modules have been explored as both affinity tags for purifying proteins and in the immobilization of industrially relevant enzymes to insoluble matrices [31, 33]. These modules are also deployed in the exploration of the architecture of plant cell walls [34].
Family Firsts
- First Identified
- A CBM2 module was first discovered in the cellulase CenA and the xylanase CexA from Cellulomonas fimi. These modules were identified through treatment of these enzymes with proteinases, which resulted in the loss of the C-terminal region comprising the CBM2 module, which reduced binding to cellulose and, in the case of CenA, activity against crystalline forms of the polysaccharide [1]. In the early literature CBM2 modules were referred to as cellulose binding domains or CBDs.
- First structural characterization
- The first 3D structure of a cellulose binding CBM2 module, which was determined by NMR, was the C. fimi xylanase Cex [10]. The first structure of a xylan binding CBM2 member, also elucidated by NMR, was from the C. fimi xylanase Xyn11A [14]. The first crystal structure of a member of CBM2 was that of the chitin binding module from the Pyrococcus chitinase Chi18A (see PDB ID 2cwr) [2].
References
- Gilkes NR, Warren RA, Miller RC Jr, and Kilburn DG. (1988). Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988;263(21):10401-7. | Google Books | Open Library
- Nakamura T, Mine S, Hagihara Y, Ishikawa K, Ikegami T, and Uegaki K. (2008). Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus. J Mol Biol. 2008;381(3):670-80. DOI:10.1016/j.jmb.2008.06.006 |
- Black GW, Hazlewood GP, Millward-Sadler SJ, Laurie JI, and Gilbert HJ. (1995). A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem J. 1995;307 ( Pt 1)(Pt 1):191-5. DOI:10.1042/bj3070191 |
- Hernandez-Gomez MC, Rydahl MG, Rogowski A, Morland C, Cartmell A, Crouch L, Labourel A, Fontes CM, Willats WG, Gilbert HJ, and Knox JP. (2015). Recognition of xyloglucan by the crystalline cellulose-binding site of a family 3a carbohydrate-binding module. FEBS Lett. 2015;589(18):2297-303. DOI:10.1016/j.febslet.2015.07.009 |
- Bolam DN, Ciruela A, McQueen-Mason S, Simpson P, Williamson MP, Rixon JE, Boraston A, Hazlewood GP, and Gilbert HJ. (1998). Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem J. 1998;331 ( Pt 3)(Pt 3):775-81. DOI:10.1042/bj3310775 |
- Bolam DN, Xie H, White P, Simpson PJ, Hancock SM, Williamson MP, and Gilbert HJ. (2001). Evidence for synergy between family 2b carbohydrate binding modules in Cellulomonas fimi xylanase 11A. Biochemistry. 2001;40(8):2468-77. DOI:10.1021/bi002564l |
- Creagh AL, Ong E, Jervis E, Kilburn DG, and Haynes CA. (1996). Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc Natl Acad Sci U S A. 1996;93(22):12229-34. DOI:10.1073/pnas.93.22.12229 |
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, and Knox JP. (2006). Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem. 2006;281(39):29321-9. DOI:10.1074/jbc.M605903200 |
- McLean BW, Boraston AB, Brouwer D, Sanaie N, Fyfe CA, Warren RA, Kilburn DG, and Haynes CA. (2002). Carbohydrate-binding modules recognize fine substructures of cellulose. J Biol Chem. 2002;277(52):50245-54. DOI:10.1074/jbc.M204433200 |
- Xu GY, Ong E, Gilkes NR, Kilburn DG, Muhandiram DR, Harris-Brandts M, Carver JP, Kay LE, and Harvey TS. (1995). Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34(21):6993-7009. | Google Books | Open Library
- Bray MR, Johnson PE, Gilkes NR, McIntosh LP, Kilburn DG, and Warren RA. (1996). Probing the role of tryptophan residues in a cellulose-binding domain by chemical modification. Protein Sci. 1996;5(11):2311-8. DOI:10.1002/pro.5560051117 |
- McLean BW, Bray MR, Boraston AB, Gilkes NR, Haynes CA, and Kilburn DG. (2000). Analysis of binding of the family 2a carbohydrate-binding module from Cellulomonas fimi xylanase 10A to cellulose: specificity and identification of functionally important amino acid residues. Protein Eng. 2000;13(11):801-9. DOI:10.1093/protein/13.11.801 |
- Nagy T, Simpson P, Williamson MP, Hazlewood GP, Gilbert HJ, and Orosz L. (1998). All three surface tryptophans in Type IIa cellulose binding domains play a pivotal role in binding both soluble and insoluble ligands. FEBS Lett. 1998;429(3):312-6. DOI:10.1016/s0014-5793(98)00625-5 |
- Simpson PJ, Bolam DN, Cooper A, Ciruela A, Hazlewood GP, Gilbert HJ, and Williamson MP. (1999). A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity. Structure. 1999;7(7):853-64. DOI:10.1016/s0969-2126(99)80108-7 |
- Simpson PJ, Xie H, Bolam DN, Gilbert HJ, and Williamson MP. (2000). The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J Biol Chem. 2000;275(52):41137-42. DOI:10.1074/jbc.M006948200 |
- Xie H, Bolam DN, Nagy T, Szabó L, Cooper A, Simpson PJ, Lakey JH, Williamson MP, and Gilbert HJ. (2001). Role of hydrogen bonding in the interaction between a xylan binding module and xylan. Biochemistry. 2001;40(19):5700-7. DOI:10.1021/bi010034z |
- Kellett LE, Poole DM, Ferreira LM, Durrant AJ, Hazlewood GP, and Gilbert HJ. (1990). Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990;272(2):369-76. DOI:10.1042/bj2720369 |
- Ferreira LM, Wood TM, Williamson G, Faulds C, Hazlewood GP, Black GW, and Gilbert HJ. (1993). A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993;294 ( Pt 2)(Pt 2):349-55. DOI:10.1042/bj2940349 |
- Brown IE, Mallen MH, Charnock SJ, Davies GJ, and Black GW. (2001). Pectate lyase 10A from Pseudomonas cellulosa is a modular enzyme containing a family 2a carbohydrate-binding module. Biochem J. 2001;355(Pt 1):155-65. DOI:10.1042/0264-6021:3550155 |
- Hogg D, Pell G, Dupree P, Goubet F, Martín-Orúe SM, Armand S, and Gilbert HJ. (2003). The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem J. 2003;371(Pt 3):1027-43. DOI:10.1042/BJ20021860 |
- Crouch LI, Labourel A, Walton PH, Davies GJ, and Gilbert HJ. (2016). The Contribution of Non-catalytic Carbohydrate Binding Modules to the Activity of Lytic Polysaccharide Monooxygenases. J Biol Chem. 2016;291(14):7439-49. DOI:10.1074/jbc.M115.702365 |
- Forsberg Z, Mackenzie AK, Sørlie M, Røhr ÅK, Helland R, Arvai AS, Vaaje-Kolstad G, and Eijsink VG. (2014). Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc Natl Acad Sci U S A. 2014;111(23):8446-51. DOI:10.1073/pnas.1402771111 |
- Bolam DN, Xie H, Pell G, Hogg D, Galbraith G, Henrissat B, and Gilbert HJ. (2004). X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J Biol Chem. 2004;279(22):22953-63. DOI:10.1074/jbc.M313317200 |
- DeBoy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB, Mohamoud Y, Watkins K, Henrissat B, Gilbert HJ, and Nelson KE. (2008). Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J Bacteriol. 2008;190(15):5455-63. DOI:10.1128/JB.01701-07 |
- Attia M, Stepper J, Davies GJ, and Brumer H. (2016). Functional and structural characterization of a potent GH74 endo-xyloglucanase from the soil saprophyte Cellvibrio japonicus unravels the first step of xyloglucan degradation. FEBS J. 2016;283(9):1701-19. DOI:10.1111/febs.13696 |
- Weiner RM, Taylor LE 2nd, Henrissat B, Hauser L, Land M, Coutinho PM, Rancurel C, Saunders EH, Longmire AG, Zhang H, Bayer EA, Gilbert HJ, Larimer F, Zhulin IB, Ekborg NA, Lamed R, Richardson PM, Borovok I, and Hutcheson S. (2008). Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T. PLoS Genet. 2008;4(5):e1000087. DOI:10.1371/journal.pgen.1000087 |
- Ferreira LM, Durrant AJ, Hall J, Hazlewood GP, and Gilbert HJ. (1990). Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990;269(1):261-4. DOI:10.1042/bj2690261 |
- Shen H, Schmuck M, Pilz I, Gilkes NR, Kilburn DG, Miller RC Jr, and Warren RA. (1991). Deletion of the linker connecting the catalytic and cellulose-binding domains of endoglucanase A (CenA) of Cellulomonas fimi alters its conformation and catalytic activity. J Biol Chem. 1991;266(17):11335-40. | Google Books | Open Library
- Din N, Damude HG, Gilkes NR, Miller RC Jr, Warren RA, and Kilburn DG. (1994). C1-Cx revisited: intramolecular synergism in a cellulase. Proc Natl Acad Sci U S A. 1994;91(24):11383-7. DOI:10.1073/pnas.91.24.11383 |
- Hervé C, Rogowski A, Blake AW, Marcus SE, Gilbert HJ, and Knox JP. (2010). Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A. 2010;107(34):15293-8. DOI:10.1073/pnas.1005732107 |
- Tomme P, Boraston A, McLean B, Kormos J, Creagh AL, Sturch K, Gilkes NR, Haynes CA, Warren RA, and Kilburn DG. (1998). Characterization and affinity applications of cellulose-binding domains. J Chromatogr B Biomed Sci Appl. 1998;715(1):283-96. DOI:10.1016/s0378-4347(98)00053-x |
- Jervis EJ, Haynes CA, and Kilburn DG. (1997). Surface diffusion of cellulases and their isolated binding domains on cellulose. J Biol Chem. 1997;272(38):24016-23. DOI:10.1074/jbc.272.38.24016 |
- Boraston AB, McLean BW, Guarna MM, Amandaron-Akow E, and Kilburn DG. (2001). A family 2a carbohydrate-binding module suitable as an affinity tag for proteins produced in Pichia pastoris. Protein Expr Purif. 2001;21(3):417-23. DOI:10.1006/prep.2001.1393 |
- McCartney L, Blake AW, Flint J, Bolam DN, Boraston AB, Gilbert HJ, and Knox JP. (2006). Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc Natl Acad Sci U S A. 2006;103(12):4765-70. DOI:10.1073/pnas.0508887103 |