CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 82"
Harry Brumer (talk | contribs) (added Etienne Rebuffet as co-author) |
|||
| Line 26: | Line 26: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | The two known members of [[glycoside hydrolase]] family 82 enzymes cleave the β-1,4 galactosidic bond of the marine algal polysaccharide iota-carrageenan <cite> | + | The two known members of [[glycoside hydrolase]] family 82 enzymes cleave the β-1,4 galactosidic bond of the marine algal polysaccharide iota-carrageenan <cite>Barbeyron2000</cite> yielding products of the neocarrabiose series. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | Family 82 enzymes are [[inverting]] enzymes, as first shown by NMR <cite> | + | Family 82 enzymes are [[inverting]] enzymes, as first shown by NMR <cite>Barbeyron2000</cite> on the iota-carrageenase from ''Alteromonas fortis''. |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | From structural analysis the catalytic residues have been predicted to be two out of the three candidate amino acids Glu245, Asp247 or Glu310 in the ''A. fortis'' iota-carrageenase <cite> | + | From structural analysis the catalytic residues have been predicted to be two out of the three candidate amino acids Glu245, Asp247 or Glu310 in the ''A. fortis'' iota-carrageenase <cite>Michel2001</cite>. A study in 2010, which utilized site directed mutagenesis, has confirmed that Glu245 plays the role of the [[general acid]] residue in this [[inverting]] enzyme, while Asp247 is the [[general base]] activating the nucleophilic water molecule <cite>Rebuffet2010</cite>. However, intriguingly the position of equivalent residues to Asp247 in other iota-carrageenase sequences are not strictly conserved <cite>Rebuffet2010 Hatada2010</cite>. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
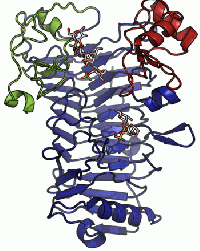
| − | A crystal structure has only been determined for the iota-carrageenase from ''A. fortis'' <cite>2</cite>. The crystal structure of a product complex has shed light on the existance of domain movement of domain A that is closed around the oligo-carrageenan in the complexed form and open in the uncomplexed enzyme <cite> | + | [[Image:gh82_domA.gif|thumb|200px|right]] |
| + | A crystal structure has only been determined for the iota-carrageenase from ''A. fortis'' <cite>2</cite>. The crystal structure of a product complex has shed light on the existance of domain movement of domain A that is closed around the oligo-carrageenan in the complexed form and open in the uncomplexed enzyme (Figure 1) <cite>Michel2003</cite>. | ||
== Family Firsts == | == Family Firsts == | ||
| − | ;First sequence identification and family creation: iota-carrageenase sequences have been first reported for enzymes from ''A. fortis'' and ''Z. galactanivorans'' <cite> | + | ;First sequence identification and family creation: iota-carrageenase sequences have been first reported for enzymes from ''A. fortis'' and ''Z. galactanivorans'' <cite>Barbeyron2000</cite>. |
| − | ;First sterochemistry determination: GH82 enzymes are inverting as shown by NMR <cite> | + | ;First sterochemistry determination: GH82 enzymes are inverting as shown by NMR <cite>Barbeyron2000</cite>. |
| − | ;First general acid residue identification: Glu245 <cite> | + | ;First general acid residue identification: Glu245 <cite>Rebuffet2010</cite> |
| − | ;First general base residue identification: Asp247 <cite> | + | ;First general base residue identification: Asp247 <cite>Rebuffet2010</cite>. |
| − | ;First 3-D structure: iota-carrageenase from ''A. fortis'' <cite> | + | ;First 3-D structure: iota-carrageenase from ''A. fortis'' <cite>Michel2001</cite>. The structure belongs to the β-helix fold ([{{PDBlink}}1h80 PDB 1h80] and [{{PDBlink}}1ktw PDB 1ktw]). |
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Barbeyron2000 pmid=10934194 |
| − | # | + | #Michel2001 pmid=11493601 |
| − | # | + | #Rebuffet2010 pmid=20681629 |
| − | # | + | #Hatada2010 pmid=20686828 |
| − | # | + | #Michel2003 pmid=14623184 |
| − | |||
</biblio> | </biblio> | ||
[[Category:Glycoside Hydrolase Families|GH082]] | [[Category:Glycoside Hydrolase Families|GH082]] | ||
Revision as of 05:57, 5 May 2011
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Authors: ^^^Mirjam Czjzek^^^, ^^^Gurvan Michel^^^, and ^^^Etienne Rebuffet^^^
- Responsible Curator: ^^^Mirjam Czjzek^^^
| Glycoside Hydrolase Family GH82 | |
| Clan | none |
| Mechanism | inverting |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/fam/GH82.html | |
Substrate specificities
The two known members of glycoside hydrolase family 82 enzymes cleave the β-1,4 galactosidic bond of the marine algal polysaccharide iota-carrageenan [1] yielding products of the neocarrabiose series.
Kinetics and Mechanism
Family 82 enzymes are inverting enzymes, as first shown by NMR [1] on the iota-carrageenase from Alteromonas fortis.
Catalytic Residues
From structural analysis the catalytic residues have been predicted to be two out of the three candidate amino acids Glu245, Asp247 or Glu310 in the A. fortis iota-carrageenase [2]. A study in 2010, which utilized site directed mutagenesis, has confirmed that Glu245 plays the role of the general acid residue in this inverting enzyme, while Asp247 is the general base activating the nucleophilic water molecule [3]. However, intriguingly the position of equivalent residues to Asp247 in other iota-carrageenase sequences are not strictly conserved [3, 4].
Three-dimensional structures
A crystal structure has only been determined for the iota-carrageenase from A. fortis [5]. The crystal structure of a product complex has shed light on the existance of domain movement of domain A that is closed around the oligo-carrageenan in the complexed form and open in the uncomplexed enzyme (Figure 1) [6].
Family Firsts
- First sequence identification and family creation
- iota-carrageenase sequences have been first reported for enzymes from A. fortis and Z. galactanivorans [1].
- First sterochemistry determination
- GH82 enzymes are inverting as shown by NMR [1].
- First general acid residue identification
- Glu245 [3]
- First general base residue identification
- Asp247 [3].
- First 3-D structure
- iota-carrageenase from A. fortis [2]. The structure belongs to the β-helix fold (PDB 1h80 and PDB 1ktw).
References
- Barbeyron T, Michel G, Potin P, Henrissat B, and Kloareg B. (2000). iota-Carrageenases constitute a novel family of glycoside hydrolases, unrelated to that of kappa-carrageenases. J Biol Chem. 2000;275(45):35499-505. DOI:10.1074/jbc.M003404200 |
- Michel G, Chantalat L, Fanchon E, Henrissat B, Kloareg B, and Dideberg O. (2001). The iota-carrageenase of Alteromonas fortis. A beta-helix fold-containing enzyme for the degradation of a highly polyanionic polysaccharide. J Biol Chem. 2001;276(43):40202-9. DOI:10.1074/jbc.M100670200 |
- Rebuffet E, Barbeyron T, Jeudy A, Jam M, Czjzek M, and Michel G. (2010). Identification of catalytic residues and mechanistic analysis of family GH82 iota-carrageenases. Biochemistry. 2010;49(35):7590-9. DOI:10.1021/bi1003475 |
- Hatada Y, Mizuno M, Li Z, and Ohta Y. (2011). Hyper-production and characterization of the ι-carrageenase useful for ι-carrageenan oligosaccharide production from a deep-sea bacterium, Microbulbifer thermotolerans JAMB-A94T, and insight into the unusual catalytic mechanism. Mar Biotechnol (NY). 2011;13(3):411-22. DOI:10.1007/s10126-010-9312-0 |
- Michel G, Helbert W, Kahn R, Dideberg O, and Kloareg B. (2003). The structural bases of the processive degradation of iota-carrageenan, a main cell wall polysaccharide of red algae. J Mol Biol. 2003;334(3):421-33. DOI:10.1016/j.jmb.2003.09.056 |