CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Glycoside Hydrolase Family 70"
| Line 37: | Line 37: | ||
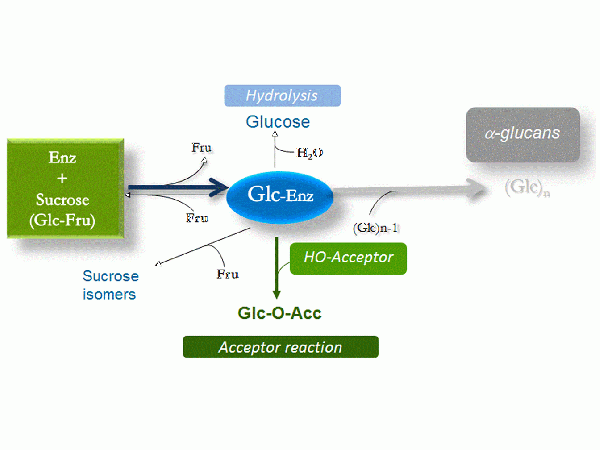
Sucrose analogues such as α-D-glucopyranosyl fluoride, ''p''-nitrophenyl α-D-glucopyranoside, α-D-glucopyranosyl α-L-sorofuranoside and lactulosucrose were also reported as D-glucopyranosyl donors <cite>Genghof1972,Figures1976,Binder1983</cite>. GH70 glucansucrases also transfer the D-glucosyl units from sucrose onto hydroxyl acceptor groups. A large variety of acceptors may be recognized by glucansucrases including carbohydrates, alcohols, polyols or flavonoids to yield oligosaccharides or gluco-conjugates <cite>Monchois1999,Leemhuis2012,Andre2010,Koepsell1953,Nakahara1995,Richard2003,Bertrand2006</cite>. The structure of the resultant glucosylated product is dependent upon the enzyme specificity. | Sucrose analogues such as α-D-glucopyranosyl fluoride, ''p''-nitrophenyl α-D-glucopyranoside, α-D-glucopyranosyl α-L-sorofuranoside and lactulosucrose were also reported as D-glucopyranosyl donors <cite>Genghof1972,Figures1976,Binder1983</cite>. GH70 glucansucrases also transfer the D-glucosyl units from sucrose onto hydroxyl acceptor groups. A large variety of acceptors may be recognized by glucansucrases including carbohydrates, alcohols, polyols or flavonoids to yield oligosaccharides or gluco-conjugates <cite>Monchois1999,Leemhuis2012,Andre2010,Koepsell1953,Nakahara1995,Richard2003,Bertrand2006</cite>. The structure of the resultant glucosylated product is dependent upon the enzyme specificity. | ||
| − | [[Image:CAZypedia_GH70_Figure_1.gif|thumb|left|Reactions catalyzed from sucrose]] | + | [[Image:CAZypedia_GH70_Figure_1.gif|thumb|left||600px|Reactions catalyzed from sucrose(''click to enlarge'')]] |
Glucansucrases also catalyze glucosyl exchange between their glucan products in a reaction known as disproportionation <cite>Binderb1983</cite>. Of note, some members of GH70 family produced by ''Lb. reuteri'' sp. were also reported to be able to use maltooligosaccharides as the glucosyl donor and to catalyze its transfer to maltooligosaccharide acceptors via the formation of α-1,6 linkages, thus acting as 4,6-α-glucanotransferases and revealing additional evolutionary relations between families GH70 and [[GH13]] <cite>Kralj2011,Dobruchowska2012 </cite>. | Glucansucrases also catalyze glucosyl exchange between their glucan products in a reaction known as disproportionation <cite>Binderb1983</cite>. Of note, some members of GH70 family produced by ''Lb. reuteri'' sp. were also reported to be able to use maltooligosaccharides as the glucosyl donor and to catalyze its transfer to maltooligosaccharide acceptors via the formation of α-1,6 linkages, thus acting as 4,6-α-glucanotransferases and revealing additional evolutionary relations between families GH70 and [[GH13]] <cite>Kralj2011,Dobruchowska2012 </cite>. | ||
Revision as of 04:00, 27 August 2012
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author:
- Responsible Curator: ^^^Magali Remaud-Simeon^^^
| Glycoside Hydrolase Family GH70 | |
| Clan | GH-H |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH70.html | |
Substrate specificities
GH70 family enzymes are transglucosylases produced by lactic acid bacteria from Streptococcus, Leuconostoc, Weisella or Lactobacillus genera. Together with the families GH13 and GH77 enzymes, they form the clan GH-H.
Most of the enzymes classified in this family use sucrose as the D-glucopyranosyl donor to synthesize α-D-glucans of high molecular mass (>106 Da) with the concomitant release of D-fructose. They are also referred to as glucosyltransferases (GTF) or glucansucrases (GS) [1, 2, 3, 4]. The first glucansucrase was isolated from sucrose-broth cultures of a strain of Ln. mesenteroides [5, 6]. The enzyme was shown to synthesize water-soluble dextran (α-1,6 linked D-glucan) from sucrose alone. Dextran from Ln. mesenteroides sp. was the first microbial polysaccharide to be commercialized and used for pharmaceutical and fine chemical applications. The glucansucrases produced by Streptococcus sp. were extensively studied on account of their major role in the formation of dental caries [1, 2].
Depending on their specificity, glucansucrases catalyze the formation of linear as well as branched α-D-glucan chains with various types of glycosidic linkages, namely α-1,2; α-1,3; α-1,4; and/or α-1,6 [1, 4, 7]. They are classified by the nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the basis of the reaction catalyzed and the specificity: dextransucrase (sucrose:1,6-α-D-glucosyltransferase; EC 2.4.1.5), alternansucrase (sucrose:1,6(1,3)-α-D-glucan-6(3)-α-D-glucosyltransferase, EC 2.4.1.140), mutansucrase (sucrose:1,3-α-D-glucan-3-α-D-glucosyltransferase; EC 2.4.1.5) and reuteransucrase (sucrose:1,4(6-α-D-glucan-4(6)-α-D-glucosyltransferase; EC 2.4.1.5). A wide range of α-D-glucans, varying in size, structure, degree of branching and spatial arrangements can thus be produced by GH70 family members [1, 2, 3, 4].
Sucrose analogues such as α-D-glucopyranosyl fluoride, p-nitrophenyl α-D-glucopyranoside, α-D-glucopyranosyl α-L-sorofuranoside and lactulosucrose were also reported as D-glucopyranosyl donors [8, 9, 10]. GH70 glucansucrases also transfer the D-glucosyl units from sucrose onto hydroxyl acceptor groups. A large variety of acceptors may be recognized by glucansucrases including carbohydrates, alcohols, polyols or flavonoids to yield oligosaccharides or gluco-conjugates [2, 3, 4, 11, 12, 13, 14]. The structure of the resultant glucosylated product is dependent upon the enzyme specificity.
Glucansucrases also catalyze glucosyl exchange between their glucan products in a reaction known as disproportionation [15]. Of note, some members of GH70 family produced by Lb. reuteri sp. were also reported to be able to use maltooligosaccharides as the glucosyl donor and to catalyze its transfer to maltooligosaccharide acceptors via the formation of α-1,6 linkages, thus acting as 4,6-α-glucanotransferases and revealing additional evolutionary relations between families GH70 and GH13 [16, 17].
One glucansucrase was shown to possess two catalytically active domains acting in tandem [18]. This enzyme was shown to be secreted by Ln. mesenteroides NRRLB-1299 (reclassified as Ln. citreum). These domains work cooperatively to synthesize dextran with more than 20% α-1,2 branched linkages and are both classified in the GH70 family. One domain is responsible for the synthesis of the α-1,6 linked dextran chain, whereas the second one is specific for the catalysis of α-1,2 branch formation. The truncated form of this enzyme carrying only the second catalytic domain was named α-1,2 branching sucrase as it does not catalyse sucrose polymerization and shows transglucosylase activity only in the presence of sucrose and linear α-1,6 dextran [19].
Kinetics and Mechanism
Enzymes from GH family 70 belong to the same GH H as clan GH family 13 and 77 enzymes. They employ an a-retaining mechanism. The transfer of the glucosyl unit proceeds through the formation of a b-glucosyl ester of an aspartate side chain, which was first isolated by sequencing peptides provided by rapid quenching of enzymic reactions with labeled sucrose and extracellular glucosyltransferases from S. sobrinus [20, 21]. Once the glucosyl-enzyme intermediate is formed, the glucosyl units originating from sucrose may be transferred onto either water (hydrolysis reaction), D-fructose released from sucrose cleavage (sucrose isomer formation) or onto the growing glucan chain (main reaction) [20, 21, 22, 23]. When acceptors are introduced into the reaction mixture, acceptor glucosylation occurs at the expense of polymer formation, the competition between the two reactions being controlled by acceptor recognition [11]. Polymer extension proceeds from the non-reducing end [22] and involves a single active site as observed in the tertiary structures of GH70 enzymes [23, 24].
The catalytic domain of family 70 enzymes is often flanked at the N- and/or C-termini by sequences containing a series of homologous repeating units that confer glucan binding ability [2, 3, 25]. They are found in domain V (see structure scheme below) and have also been shown to influence polymer size, enzyme activity, specificity and processivity [2, 3, 22]. However, their precise role at the molecular level is still not fully understood [23, 24].
Catalytic Residues
Sequence analysis, secondary structure prediction and tertiary structure comparison with enzymes from GH Family 13 enabled the localization of a putative catalytic triad that is strictly conserved in family 70 and similar to that found in GH family 13 [23, 24]. This triad is formed by an aspartate (nucleophile) corresponding to the residue identified by peptide sequencing [20, 21], a glutamate (acid/base catalyst) and a second aspartic acid that is critical for transition state stabilization [23, 24]. The putative role of these residues in catalysis was further supported by mutations and structural analyses [2, 3, 4].
Three-dimensional structures
The 3D structures of GTF180-DN dextransucrase from Lb reuteri 180 [23], GTF-SI from S. mutans [24] and a-1,2 branching sucrase (engineered from DSR-E dextransucrase from Ln. citreum NRRL B-1299) [19], all revealed a unique fold organized as five distinct domains, named C, A, B, IV and V, and made up of peptide segments that are not consecutively arranged along the peptide chain. The catalytic domain of GH70 glucansucrases consists of a (b/a)8 barrel resembling that of family 13 enzymes but formed by sequences that have undergone circular permutation compared with the GH13 family domains [23, 26]. The 3D structures of complexes with sucrose, maltose and acarbose enabled the mapping of the active site and the identification of some determinants of GH70 enzyme product specificity.
Available structures
GTF180-DN (dextransucrase from Lb reuteri 180) : 3KLK, 3KLL, 3HZ3 [23]
GTF-SI (mutansucrase from S. mutans : 3AIE, 3AIC, 3AIB [24]
GBD-CD2 (a-1,2 branching sucrase) : 3TTO [24]
Family Firsts
- First stereochemistry determination
- The stereochemistry was directly linked to the discovery of the enzyme from Ln. mesenteroides catalyzing dextran synthesis from sucrose by Hehre [5]
- First amino acid sequences
- Those of GTF-B from S. mutans and GTF-I from S. sobrinus Mfe28 simultaneously reported in two companion articles [27, 28]
- First catalytic nucleophile identification
- identified first in Streptococcus sobrinus glucansucrase [20, 21]
- First general acid/base residue identification
- On the basis of sequence alignments and secondary structure [26]. On the basis of 3D-structure [23, 29]
- First 3-D structure
- The first 3-D structure for GH70 members was that of the GTF180-DN dextransucrase from Lb reuteri [23, 29]
References
- Sidebotham RL (1974). Dextrans. Adv Carbohydr Chem Biochem. 1974;30:371-444. DOI:10.1016/s0065-2318(08)60268-1 |
- Monchois V, Willemot RM, and Monsan P. (1999). Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol Rev. 1999;23(2):131-51. DOI:10.1111/j.1574-6976.1999.tb00394.x |
- Leemhuis H, Pijning T, Dobruchowska JM, van Leeuwen SS, Kralj S, Dijkstra BW, and Dijkhuizen L. (2013). Glucansucrases: three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J Biotechnol. 2013;163(2):250-72. DOI:10.1016/j.jbiotec.2012.06.037 |
- André I, Potocki-Véronèse G, Morel S, Monsan P, and Remaud-Siméon M. (2010). Sucrose-utilizing transglucosidases for biocatalysis. Top Curr Chem. 2010;294:25-48. DOI:10.1007/128_2010_52 |
- Hehre EJ (1941). PRODUCTION FROM SUCROSE OF A-SERO-LOGICALLY REACTIVE POLYSACCHARIDE BY A STERILE BACTERIAL EXTRACT. Science. 1941;93(2410):237-8. DOI:10.1126/science.93.2410.237 |
- Hehre EJ and Sugg JY. (1942). SEROLOGICALLY REACTIVE POLYSACCHARIDES PRODUCED THROUGH THE ACTION OF BACTERIAL ENZYMES : I. DEXTRAN OF LEUCONOSTOC MESENTEROIDES FROM SUCROSE. J Exp Med. 1942;75(3):339-53. DOI:10.1084/jem.75.3.339 |
-
Jeanes A, Haynes WC, Wilham CA, Rankin JC, Melvin EH, Austin MJ, Cluskey JE,Fisher BE, Tsuchiya HM, Rist CE Characterization and classification of dextrans from 96 strains of bacteria. J Am Chem Soc 1954; 76(20) 5041-5052. Doi: 10.1021/ja01649a011
- Genghof DS and Hehre EJ. (1972). De novo glycosidic linkage synthesis by glycosylases: alpha-D-glucosyl fluoride polymerization by dextransucrase. 1. Proc Soc Exp Biol Med. 1972;140(4):1298-301. DOI:10.3181/00379727-140-36662 |
- Figures WR and Edwards JR. (1976). alpha-D-Glucopyranosyl fluoride as a D-glucopyranosyl donor for a glycosyltransferase complex from Streptococcus mutans FA1. Carbohydr Res. 1976;48(2):245-53. DOI:10.1016/s0008-6215(00)83220-6 |
- Binder TP and Robyt JF. (1983). p-Nitrophenyl alpha-D-glucopyranoside, a new substrate for glucansucrases. Carbohydr Res. 1983;124(2):287-99. DOI:10.1016/0008-6215(83)88464-x |
- KOEPSELL HJ, TSUCHIYA HM, HELLMAN NN, KAZENKO A, HOFFMAN CA, SHARPE ES, and JACKSON RW. (1953). Enzymatic synthesis of dextran; acceptor specificity and chain initiation. J Biol Chem. 1953;200(2):793-801. | Google Books | Open Library
- Nakahara K, Kontani M, Ono H, Kodama T, Tanaka T, Ooshima T, and Hamada S. (1995). Glucosyltransferase from Streptococcus sobrinus Catalyzes Glucosylation of Catechin. Appl Environ Microbiol. 1995;61(7):2768-70. DOI:10.1128/aem.61.7.2768-2770.1995 |
- Richard G, Morel S, Willemot RM, Monsan P, and Remaud-Simeon M. (2003). Glucosylation of alpha-butyl- and alpha-octyl-D-glucopyranosides by dextransucrase and alternansucrase from Leuconostoc mesenteroides. Carbohydr Res. 2003;338(9):855-64. DOI:10.1016/s0008-6215(03)00070-3 |
- Bertrand A, Morel S, Lefoulon F, Rolland Y, Monsan P, and Remaud-Simeon M. (2006). Leuconostoc mesenteroides glucansucrase synthesis of flavonoid glucosides by acceptor reactions in aqueous-organic solvents. Carbohydr Res. 2006;341(7):855-63. DOI:10.1016/j.carres.2006.02.008 |
- Binder TP, Côté GL, and Robyt JF. (1983). Disproportionation reactions catalyzed by Leuconostoc and Streptococcus glucansucrases. Carbohydr Res. 1983;124(2):275-86. DOI:10.1016/0008-6215(83)88463-8 |
- Kralj S, Grijpstra P, van Leeuwen SS, Leemhuis H, Dobruchowska JM, van der Kaaij RM, Malik A, Oetari A, Kamerling JP, and Dijkhuizen L. (2011). 4,6-α-glucanotransferase, a novel enzyme that structurally and functionally provides an evolutionary link between glycoside hydrolase enzyme families 13 and 70. Appl Environ Microbiol. 2011;77(22):8154-63. DOI:10.1128/AEM.05735-11 |
- Dobruchowska JM, Gerwig GJ, Kralj S, Grijpstra P, Leemhuis H, Dijkhuizen L, and Kamerling JP. (2012). Structural characterization of linear isomalto-/malto-oligomer products synthesized by the novel GTFB 4,6-α-glucanotransferase enzyme from Lactobacillus reuteri 121. Glycobiology. 2012;22(4):517-28. DOI:10.1093/glycob/cwr167 |
- Fabre E, Bozonnet S, Arcache A, Willemot RM, Vignon M, Monsan P, and Remaud-Simeon M. (2005). Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly alpha-1,2 branched dextran. J Bacteriol. 2005;187(1):296-303. DOI:10.1128/JB.187.1.296-303.2005 |
- Brison Y, Pijning T, Malbert Y, Fabre É, Mourey L, Morel S, Potocki-Véronèse G, Monsan P, Tranier S, Remaud-Siméon M, and Dijkstra BW. (2012). Functional and structural characterization of α-(1->2) branching sucrase derived from DSR-E glucansucrase. J Biol Chem. 2012;287(11):7915-24. DOI:10.1074/jbc.M111.305078 |
- Mooser G and Iwaoka KR. (1989). Sucrose 6-alpha-D-glucosyltransferase from Streptococcus sobrinus: characterization of a glucosyl-enzyme complex. Biochemistry. 1989;28(2):443-9. DOI:10.1021/bi00428a006 |
- Mooser G, Hefta SA, Paxton RJ, Shively JE, and Lee TD. (1991). Isolation and sequence of an active-site peptide containing a catalytic aspartic acid from two Streptococcus sobrinus alpha-glucosyltransferases. J Biol Chem. 1991;266(14):8916-22. | Google Books | Open Library
- Moulis C, Joucla G, Harrison D, Fabre E, Potocki-Veronese G, Monsan P, and Remaud-Simeon M. (2006). Understanding the polymerization mechanism of glycoside-hydrolase family 70 glucansucrases. J Biol Chem. 2006;281(42):31254-67. DOI:10.1074/jbc.M604850200 |
- Vujicić-Zagar A and Dijkstra BW. (2006). Monoclinic crystal form of Aspergillus niger alpha-amylase in complex with maltose at 1.8 angstroms resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62(Pt 8):716-21. DOI:10.1107/S1744309106024729 |
- Ito K, Ito S, Shimamura T, Weyand S, Kawarasaki Y, Misaka T, Abe K, Kobayashi T, Cameron AD, and Iwata S. (2011). Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J Mol Biol. 2011;408(2):177-86. DOI:10.1016/j.jmb.2011.02.028 |
- Janecek S, Svensson B, and Russell RR. (2000). Location of repeat elements in glucansucrases of Leuconostoc and Streptococcus species. FEMS Microbiol Lett. 2000;192(1):53-7. DOI:10.1111/j.1574-6968.2000.tb09358.x |
- MacGregor EA, Jespersen HM, and Svensson B. (1996). A circularly permuted alpha-amylase-type alpha/beta-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 1996;378(3):263-6. DOI:10.1016/0014-5793(95)01428-4 |
- Shiroza T, Ueda S, and Kuramitsu HK. (1987). Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol. 1987;169(9):4263-70. DOI:10.1128/jb.169.9.4263-4270.1987 |
- Ferretti JJ, Gilpin ML, and Russell RR. (1987). Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J Bacteriol. 1987;169(9):4271-8. DOI:10.1128/jb.169.9.4271-4278.1987 |
-
Pijning T, Vujicic-Zagar A, Kralj S, Eeuwema W, Dijkhuizen L, Dijkstra, BW Biochemical and crystallographic characterization of a glucansucrase from Lactobacillus reuteri 180. Biocat Biotrans 2008; 26 (1-2) 12-17 DOI: 10.1080/10242420701789163