CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 117
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Etienne Rebuffet^^^
- Responsible Curator: ^^^Mirjam Czjzek^^^
| Glycoside Hydrolase Family GH117 | |
| Clan | None |
| Mechanism | Not known |
| Active site residues | Not known |
| CAZy DB link | |
| https://www.cazy.org/GH117.html | |
Substrate specificities
The only activity so far characterized within this recently discovered family of glycoside hydrolases is that of α-1,3-L-(3,6-anhydro)-galactosidase [1, 2, 3, 4, 5]. Nevertheless phylogenetic analyses (Figure 1) of this family together with activity tests for another member, Zg3597 (Clade C), show that the family GH117 most probably is polyspecific [3].

Kinetics and Mechanism
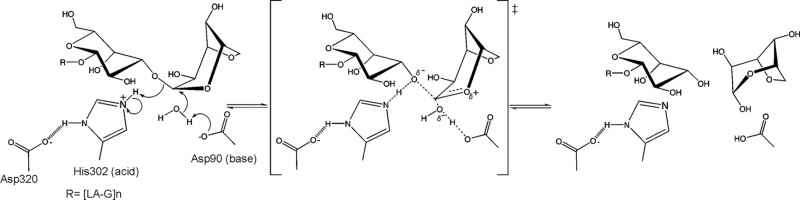
The stereochemical outcome of members of glycoside hydrolase family GH117 is still not determined experimentally. Nevertheless a mechanism based on the structure of an inactive mutant (BpGH117 E303Q) complexed to a neoagarobiose has been proposed [5] (Figure 2). In this unusual inverting catalytic mechanism an aspartic acid acting as the base and a histidine acting as the acid. An analogous Asp-His dyad has been similarly reported to act as the general base catalyst in the retaining mechanism of select GH3 members [6].
Two of the three 3D structures revealed the presence of a divalent cation, directly coordinated only by water molecules, close to the active site, which could activate the catalytic water molecule and provide the energy needed for the enzymatic reaction [3, 5]. Sequence alignments suggest that the enzymes of clades B and C do not bind divalent cation, which could be related to their difference in substrate specificity [3].
Catalytic Residues
From structural analysis and sequence alignments the catalytic residues have been predicted to be Asp-90 as the base and His-302 as the acid BpGH117 numbering) [5].
Three-dimensional structures
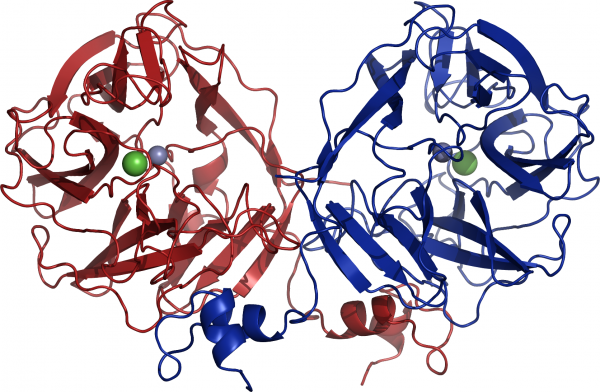
Three crystal structures of GH117 family have been reported. Two are enzymes from marine bacteria, one from Saccharophagus degradans (PDB: 3R4Y) [4] and one from Zobellia galactanivorans (PDB: 3P2N) [3], the third one is from the human gut bacterium Bacteroidetes plebeius (PDB: 4AK5) [5]. GH117 adopts a five-bladed β-propeller fold and forms a dimer via domain-swapping of the N-terminal HTH (Helix-Turn-Helix) domain (Figure 3) [3]. Interestingly, previous sequences reported from Vibrio sp. JT0107 and Bacillus sp. MK03 contain the conserved domain-swapping signature SxAxxR in the HTH domain. Consistently, these proteins were reported to form multimers (a dimer and an octamer respectively), based on calibrated gel filtration estimations [1, 2]. In contrast, RB13146 (Clade B) lacks the domain-swapping signature, in which the crucial residues are missing. This enzyme from R. baltica thus likely occurs as a monomer and may represent an ‘ancestral’ form of the GH117 family, which would be limited to the catalytic β-propeller domain [3]. Structure of SdNABH and BpGH117 possess a ordered C terminus part which also interact with the adjacent monomer [4, 5]. Moreover in the case of BpGH117, His-392 from the C terminus of the monomer A participate in the substrate binding in the binding pocket of monomer B, and aims versa [5].
Family Firsts
- First stereochemistry determination
- not determined yet.
- First catalytic nucleophile identification
- not determined yet.
- First general acid/base residue identification
- not determined yet.
- First 3-D structure
- The first 3D structure was reported in 2011 for an α-1,3-L-(3,6-anhydro)-galactosidase (AhgA or Zg4663) from the marine bacteria Zobellia galactanivorans, PDB: 3p2n [3].
References
- Sugano Y, Kodama H, Terada I, Yamazaki Y, and Noma M. (1994). Purification and characterization of a novel enzyme, alpha-neoagarooligosaccharide hydrolase (alpha-NAOS hydrolase), from a marine bacterium, Vibrio sp. strain JT0107. J Bacteriol. 1994;176(22):6812-8. DOI:10.1128/jb.176.22.6812-6818.1994 |
- Suzuki H, Sawai Y, Suzuki T, and Kawai K. (2002). Purification and characterization of an extracellular alpha-neoagarooligosaccharide hydrolase from Bacillus sp. MK03. J Biosci Bioeng. 2002;93(5):456-63. DOI:10.1016/s1389-1723(02)80092-5 |
- Rebuffet E, Groisillier A, Thompson A, Jeudy A, Barbeyron T, Czjzek M, and Michel G. (2011). Discovery and structural characterization of a novel glycosidase family of marine origin. Environ Microbiol. 2011;13(5):1253-70. DOI:10.1111/j.1462-2920.2011.02426.x |
- Ha SC, Lee S, Lee J, Kim HT, Ko HJ, Kim KH, and Choi IG. (2011). Crystal structure of a key enzyme in the agarolytic pathway, α-neoagarobiose hydrolase from Saccharophagus degradans 2-40. Biochem Biophys Res Commun. 2011;412(2):238-44. DOI:10.1016/j.bbrc.2011.07.073 |
- Hehemann JH, Smyth L, Yadav A, Vocadlo DJ, and Boraston AB. (2012). Analysis of keystone enzyme in Agar hydrolysis provides insight into the degradation (of a polysaccharide from) red seaweeds. J Biol Chem. 2012;287(17):13985-95. DOI:10.1074/jbc.M112.345645 |
- Litzinger S, Fischer S, Polzer P, Diederichs K, Welte W, and Mayer C. (2010). Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism. J Biol Chem. 2010;285(46):35675-84. DOI:10.1074/jbc.M110.131037 |