CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 16"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) |
||
| Line 76: | Line 76: | ||
== Evolution of GH16 == | == Evolution of GH16 == | ||
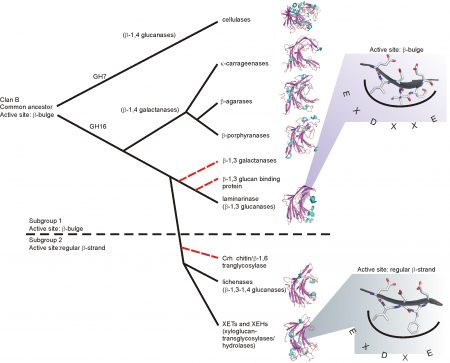
[[Image:TreeGH16new.png|thumb|right|450px|'''Figure 1. Proposed evolution of GH16 (''click to enlarge'').''']] | [[Image:TreeGH16new.png|thumb|right|450px|'''Figure 1. Proposed evolution of GH16 (''click to enlarge'').''']] | ||
| − | GH16 is a member of [[clans|clan]] GH-B together with [[GH7]]; both families share the β-jellyroll fold. The different specificities of GH16 are proposed to have evolved from an ancestral β-1,3-glucanase <cite>Barbeyron1998</cite>. This proposal was elaborated using a structure-based phylogeny approach, which suggested that a first branching event lead to the evolution of the bacterial κ-carrageenases and the β-agarases, while a later branching event lead to the bacterial licheninases and the plant XETs <cite>Michel2001</cite> (Figure 1). In particular, the GH16 active-site residues are located in-train on one beta-strand at the center of the substrate binding cleft. Depending upon the phylogenetic clade, this beta-strand features one of two topologies. The beta-bulge motif, which has the consensus sequence EXDXXE, is more frequent in GH16 compared to the regular beta-strand with the consensus sequence EXDXE (the [[catalytic nucleophile]] is the first glutamate and the [[catalytic acid/base]] is the second, with a proposed "helper" asparate in-between <cite>Planas2000</cite>). Due to the predominance of the beta-bulge motif and its presence as the only motif in [[GH7]], Michel et al. proposed that the beta-bulge is the ancestral motif, which subsequently gave rise to the regular beta-strand of extant XETs and licheninases <cite>Michel2001</cite>. More recently, similar structure-based phylogenetic approaches have suggested that XEHs evolved subsequently to XEHs within the ''xyloglucan endo-transglycosylase/hydrolase (XTH)'' gene family in plant lineages <cite>Baumann2007 Eklof2010</cite>, while the recent identification of a group of bifunctional GH16 [[glycoside hydrolases]], which is active on both mixed-linkage beta-glucan and xyloglucan, provides additional support for the close evolutionary relationship of XETs and licheninases <cite>Eklof2013</cite>. | + | GH16 is a member of [[clans|clan]] GH-B together with [[GH7]]; both families share the β-jellyroll fold. The different specificities of GH16 are proposed to have evolved from an ancestral β-1,3-glucanase <cite>Barbeyron1998</cite>. This proposal was elaborated using a structure-based phylogeny approach, which suggested that a first branching event lead to the evolution of the bacterial κ-carrageenases and the β-agarases, while a later branching event lead to the bacterial licheninases and the plant XETs <cite>Michel2001</cite> (Figure 1). In particular, the GH16 active-site residues are located in-train on one beta-strand at the center of the substrate binding cleft. Depending upon the phylogenetic clade, this beta-strand features one of two topologies. The beta-bulge motif, which has the consensus sequence EXDXXE, is more frequent in GH16 compared to the regular beta-strand with the consensus sequence EXDXE (the [[catalytic nucleophile]] is the first glutamate and the [[catalytic acid/base]] is the second, with a proposed "helper" asparate in-between <cite>Planas2000</cite>). Due to the predominance of the beta-bulge motif and its presence as the only motif in [[GH7]], Michel et al. proposed that the beta-bulge is the ancestral motif, which subsequently gave rise to the regular beta-strand of extant XETs and licheninases <cite>Michel2001</cite>. More recently, similar structure-based phylogenetic approaches have suggested that XEHs evolved subsequently to XEHs within the ''xyloglucan endo-transglycosylase/hydrolase (XTH)'' gene family in plant lineages <cite>Baumann2007 Eklof2010</cite>, while the recent identification of a group of bifunctional GH16 [[glycoside hydrolases]], which is active on both mixed-linkage beta-glucan and xyloglucan, provides additional support for the close evolutionary relationship of XETs and licheninases <cite>Eklof2013 McGregor2016</cite>. |
== Family firsts == | == Family firsts == | ||
| Line 109: | Line 109: | ||
#Eklof2013 pmid=23572521 | #Eklof2013 pmid=23572521 | ||
#Bissaro2015 pmid=25793417 | #Bissaro2015 pmid=25793417 | ||
| − | + | #McGregor2016 pmid=27859885 | |
</biblio> | </biblio> | ||
<!-- DO NOT REMOVE THIS CATEGORY TAG! --> | <!-- DO NOT REMOVE THIS CATEGORY TAG! --> | ||
[[Category:Glycoside Hydrolase Families|GH016]] | [[Category:Glycoside Hydrolase Families|GH016]] | ||
Revision as of 19:51, 22 November 2016
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: Jens Eklöf and ^^^Jan-Hendrik Hehemann^^^
- Responsible Curator: ^^^Harry Brumer^^^
| Glycoside Hydrolase Family 16 | |
| Clan | GH-B |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH16.html | |
Substrate specificities
The members of family 16 are active on β-1,4 or β-1,3 glycosidic bonds in various glucans and galactans. A wide diversity of glycoside hydrolases active on plant and marine polysaccharides are found in GH16, including:
- keratan-sulfate endo-1,4-β-galactosidases (EC 3.2.1.103),
- endo-1,3-β-galactanases (EC 3.2.1.-),
- endo-1,3-β-glucanases (EC 3.2.1.39),
- endo-1,3(4)-β-glucanases (EC 3.2.1.6),
- licheninases (EC 3.2.1.73),
- β-agarases (EC 3.2.1.81),
- β-porphyranases (EC 3.2.1.178) [1],
- κ-carrageenases (EC 3.2.1.83), and
- endo-xyloglucanases (EC 3.2.1.151, a.k.a. xyloglucan endo-hydrolases, XEHs, in plants [2]).
Notably, some members of GH16 are predominant transglycosylases. These include the plant xyloglucan:xyloglucosyltransferases (EC 2.4.1.207, a.k.a. xyloglucan endo-transglycosylases, XETs) [2] and yeast chitin/beta-glucan crosslinking enzymes Crh1 and Crh2 [3, 4, 5].
Some invertebrate GH16 proteins have lost their catalytic amino acids and are involved in immune response activation through the Toll pathway upon binding of β-1,3 glucan. The role of the GH16 domain in this immune response has not been fully elucidated [6].
- Diverse polysaccharides cleaved by GH16 enzymes
Kinetics and Mechanism
Members of GH16 enzymes are retaining enzymes, as first shown by NMR [7] on an endo-1,3-1,4-β-D-glucan 4-glucanohydrolase from Bacillus licheniformis. As such, they utilize a covalent glycosyl-enzyme intermediate, which is broken-down by glycosyl transfer [8, 9] to water or a carbohydrate acceptor substrate in glycoside hydrolases or transglycosylases, respectively.
Catalytic Residues
The catalytic nucleophile of GH16 enzymes was first proposed using a non-specific epoxyalkyl β-glycoside inhibitor and identification of the site of covalent labelling using ESI-MS and Edman degradation on an endo-1,3-1,4-β-D-glucan 4-glucanohydrolase from Bacillus amyloliquefaciens [10]. This was subsequently verified by azide rescue of the E134A mutant of a Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase resulting in an α-glycosyl azide from the β-glycoside substrate [11]. The general acid/base residue was identified by making the E138A site-directed mutant of the Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase together with kinetic analysis and azide rescue, which resulted in a β-glycosyl azide product [11]. These structurally conserved catalytic residues have been confirmed in a number of other GH16 members, including plant XETs and XEHs [12, 13], and yeast Crh1 and Crh2 [5].
The mechanistic analysis of bacterial mixed-linkage endo-glucanases has been expertly reviewed in the broader context of GH16 [14].
Three-dimensional structures
Proteins in GH16 share a β-jelly-roll fold in which two β-sheets align in a curved, sandwich-like manner and present a cleft-shaped active-site bounded by loops extending from the β-strands. The first solved 3D structure was a hybrid protein of licheninase M from Paenibacillus macerans and BglA from Bacillus amyloliquefaciens (PDB 1byh) in 1992 [15]. Many three-dimensional structures have been solved of family 16 members of archeal, bacterial, and eukaryotic origin (see http://www.cazy.org/GH16_structure.html for an updated list). Of these, the first eukaryotic 3D structure was the xyloglucan endo-transglycosylase PttXET16-34 from Populus tremula×tremuloides (PDB 1umz) [16] and the first archeal 3D structure was a endo-1,3-β-glucanase Lam16 from Pyrococcus furiosus (PDB 2vy0) [17].
Evolution of GH16
GH16 is a member of clan GH-B together with GH7; both families share the β-jellyroll fold. The different specificities of GH16 are proposed to have evolved from an ancestral β-1,3-glucanase [18]. This proposal was elaborated using a structure-based phylogeny approach, which suggested that a first branching event lead to the evolution of the bacterial κ-carrageenases and the β-agarases, while a later branching event lead to the bacterial licheninases and the plant XETs [19] (Figure 1). In particular, the GH16 active-site residues are located in-train on one beta-strand at the center of the substrate binding cleft. Depending upon the phylogenetic clade, this beta-strand features one of two topologies. The beta-bulge motif, which has the consensus sequence EXDXXE, is more frequent in GH16 compared to the regular beta-strand with the consensus sequence EXDXE (the catalytic nucleophile is the first glutamate and the catalytic acid/base is the second, with a proposed "helper" asparate in-between [14]). Due to the predominance of the beta-bulge motif and its presence as the only motif in GH7, Michel et al. proposed that the beta-bulge is the ancestral motif, which subsequently gave rise to the regular beta-strand of extant XETs and licheninases [19]. More recently, similar structure-based phylogenetic approaches have suggested that XEHs evolved subsequently to XEHs within the xyloglucan endo-transglycosylase/hydrolase (XTH) gene family in plant lineages [2, 20], while the recent identification of a group of bifunctional GH16 glycoside hydrolases, which is active on both mixed-linkage beta-glucan and xyloglucan, provides additional support for the close evolutionary relationship of XETs and licheninases [21, 22].
Family firsts
- First stereochemistry determination
- Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase by NMR [7].
- First catalytic nucleophile identification
- Suggested in Bacillus amyloliquefaciens 1,3-1,4-β-D-glucan 4-glucanohydrolase via non-specific epoxyalkyl β-glycoside labeling [10]. Later verified by azide rescue of inactivated mutants [11].
- First general acid/base residue identification
- Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase, first suggested by sequence homology and mutational studies [23]. This was later verified by azide rescue of inactivated mutants [11].
- First 3-D structure
- A hybrid licheninase (Bacillus amyloliquefaciens and Paenibacillus macerans) by X-ray crystallography (PDB 1byh) [15].
Reference list
Error fetching PMID 1360982:
Error fetching PMID 8280073:
Error fetching PMID 8099449:
Error fetching PMID 8182059:
Error fetching PMID 9698381:
Error fetching PMID 19154353:
Error fetching PMID 11435116:
Error fetching PMID 9580981:
Error fetching PMID 17557806:
Error fetching PMID 11150614:
Error fetching PMID 20376150:
Error fetching PMID 21653698:
Error fetching PMID 19712587:
Error fetching PMID 18694928:
Error fetching PMID 23919454:
Error fetching PMID 25495733:
Error fetching PMID 19419143:
Error fetching PMID 18043802:
Error fetching PMID 23572521:
Error fetching PMID 25793417:
Error fetching PMID 27859885:
- Error fetching PMID 20376150:
- Eklöf JM and Brumer H. (2010). The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010;153(2):456-66. DOI:10.1104/pp.110.156844 |
- Error fetching PMID 18694928:
- Error fetching PMID 23919454:
- Error fetching PMID 25495733:
- Error fetching PMID 19712587:
- Error fetching PMID 8280073:
-
Sinnott, M.L. (1990) Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev. 90, 1171-1202. DOI: 10.1021/cr00105a006
- Error fetching PMID 25793417:
- Error fetching PMID 1360982:
- Error fetching PMID 9698381:
- Error fetching PMID 19419143:
- Error fetching PMID 18043802:
- Error fetching PMID 11150614:
- Error fetching PMID 8099449:
- Error fetching PMID 15020748:
- Error fetching PMID 19154353:
- Error fetching PMID 9580981:
- Error fetching PMID 11435116:
- Error fetching PMID 17557806:
- Error fetching PMID 23572521:
- Error fetching PMID 27859885:
- Error fetching PMID 8182059:
- Error fetching PMID 21653698: