CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 158
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH158 | |
| Clan | GH-A |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH158.html | |
Substrate specificities
Members of family 158 have been shown to display activity towards β(1,3)-glucans, making this the fourth clan GH-A glycoside hydrolase family known to contain β(1,3)-glucanase activity, alongside GH17, GH128, and GH148. The founding member of this family, Vvad_PD1638 from Victivallis vadensis, was shown to be active on carboxymethyl-curdlan in a high-throughput screen [1].
BuGH158 from the human gut bacterium Bacteroides uniformis was the first GH158 member to receive detailed characterization [2]. BuGH158 is an endo-β(1,3)-glucanase with high specificity towards laminarin from Laminaria digitata, a β(1,3)-glucan with single β(1,6)-glucose branches. BuGH158 is unable to tolerate more extensive branching as evidenced by poor activity towards other β(1,3)-glucans with longer, more frequent branches like laminarin from Eisenia bicyclis and yeast β-glucan [2]. The unbranched, linear β(1,3)-glucan curdlan was also not effectively hydrolyzed by BuGH158, due the poor solubility of this polysaccharide in water (Vvad_PD1638 described above was active on a curdlan that was chemically modified to increase water-solubility [1]).
Kinetics and Mechanism
As a family within clan GH-A, GH158 members were inferred to be retaining enzymes. Retention of anomeric stereochemistry was experimentally confirmed by 1H NMR on the product of hydrolysis of 2-chloro-4-nitrophenyl laminaribioside by BuGH158 [2]. Thus, GH158 members enzymes employ the classical Koshland double-displacement mechanism, which proceeds via a covalent glycosyl-enzyme intermediate.
Catalytic Residues
The catalytic nucleophile and general acid/base residues of BuGH158 were predicted by structural homology with other clan GH-A members to be E220 and E137. The catalytic importance of these residues was subsequently confirmed by site-directed mutagenesis [2]. This glutamate pair is located on loops immediately following β-strands 7 (nucleophile) and 4 (acid/base), consistent with all other clan GH-A enzymes.
Three-dimensional structures
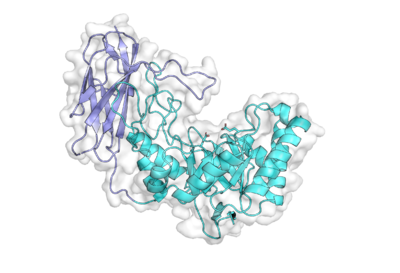
The X-ray crystal structure of BuGH158 from Bacteroides uniformis, determined by multi-wavelength anomalous dispersion, represents the founding structural representative of this family [2]. The 1.8 Å-resolution structure revealed a two-domain architecture with an N-terminal (α/β)8 triose phosphate isomerase (TIM) barrel domain (the hallmark of clan GH-A structures) and a C-terminal eight-stranded immunoglobulin (Ig)-like domain that makes extensive contacts with the TIM barrel. A loop from the Ig-like domain extends over the TIM barrel to shape the active site cleft [2].
Family Firsts
- First stereochemistry determination
- Retention of product anomeric stereochemistry by BuGH158 from Bacteroides uniformis using 1H NMR [2].
- First catalytic nucleophile identification
- E220 in BuGH158 from Bacteroides uniformis by tertiary structural homology and kinetic analysis of a site-directed mutant [2].
- First general acid/base residue identification
- E137 in BuGH158 from Bacteroides uniformis by tertiary structural homology and kinetic analysis of a site-directed mutant [2].
- First 3-D structure
- BuGH158 from Bacteroides uniformis by X-ray crystallography [2].
References
- Helbert W, Poulet L, Drouillard S, Mathieu S, Loiodice M, Couturier M, Lombard V, Terrapon N, Turchetto J, Vincentelli R, and Henrissat B. (2019). Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc Natl Acad Sci U S A. 2019;116(13):6063-6068. DOI:10.1073/pnas.1815791116 |
- Déjean G, Tamura K, Cabrera A, Jain N, Pudlo NA, Pereira G, Viborg AH, Van Petegem F, Martens EC, and Brumer H. (2020). Synergy between Cell Surface Glycosidases and Glycan-Binding Proteins Dictates the Utilization of Specific Beta(1,3)-Glucans by Human Gut Bacteroides. mBio. 2020;11(2). DOI:10.1128/mBio.00095-20 |