CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Binding Module Family 55"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (10 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * [[Author]]: | + | * [[Author]]: [[User:John Samuelson|John Samuelson]] |
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Elizabeth Ficko-Blean|Elizabeth Ficko-Blean]] |
---- | ---- | ||
| Line 17: | Line 17: | ||
== Ligand specificities == | == Ligand specificities == | ||
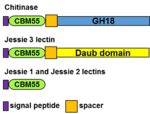
| − | A CBM55, which is ~60-aa long and contains eight cysteines in conserved positions, was first identified at the N-terminus of the [[GH18]] chitinase of ''Entamoeba histolytica'', the protist that causes dysentery and liver abscess ( | + | [[File:CBM55.png|thumb|150px|right|'''Figure 1.''' ''Entamoeba'' proteins containing CBM55.]] |
| + | A CBM55, which is ~60-aa long and contains eight cysteines in conserved positions, was first identified at the N-terminus of the [[GH18]] chitinase of ''Entamoeba histolytica'', the protist that causes dysentery and liver abscess (Figure 1) <cite>Van_Dellen2002,de_la_Vega1997</cite>. CBM55 members are also present at the N-termini of Jessie 3 lectins, which contain a self-aggregating (daub) domain, while CBM55 is the only domain in Jessie 1 and Jessie 2 lectins <cite>Chatterjee2009</cite>. CBM55 members are present in [[GH18]] chitinases and Jessie lectins of all five ''Entamoeba'' species that have been sequenced <cite>Aurrecoechea2011</cite>. CBM55 members are absent from other eukaryotes, eubacteria, and archaea, and so the CBM55 motif appears to have been “created from scratch” by the common ancestor to ''Entamoebae''. CBM55 members of ''E. histolytica'' chitinase, Jessie 1, and Jessie 3 lectins, each produced by gene expression under a constitutive actin promoter in transformed trophozoites, demonstrated binding to chitin beads <cite>Van_Dellen2002</cite>. | ||
== Structural Features == | == Structural Features == | ||
| − | While the presence of eight cysteines in conserved positions suggest that CBM55 is a disulfide knot, there is no structure for CBM55 <cite>Van_Dellen2002</cite>. Because CBM55 binds to chitin fibrils in the cyst wall, it is | + | While the presence of eight cysteines in conserved positions suggest that CBM55 is a disulfide knot, there is no structure for CBM55 <cite>Van_Dellen2002</cite>. Because CBM55 binds to chitin fibrils in the cyst wall, it is a [[Carbohydrate-binding_modules#Types|type A]] CBM <cite>Chatterjee2009</cite>. |
== Functionalities == | == Functionalities == | ||
| − | '' | + | In the human intestine, motile ''E. histolytica'' trophozoites phagocytose bacteria and/or attach to and take bites out of (trogocytose) host epithelial cells <cite>Ralston2014</cite>. Alternatively, trophozoites form cysts, which have chitin and deacetylated chitin (chitosan) in their wall <cite>Van_Dellen2006a,Das2006</cite>. ''E. histolytica'' is transmitted when cysts are shed in the feces and contaminate food, soil, or water. When cysts are ingested, trophozoites escape their wall and are released into the intestine, completing the life cycle. |
| − | + | The most abundant proteins identified by mass spectrometry of cyst walls of ''E. invadens'', a model for the human pathogen, are chitinases, Jessie 3 lectins, and Jacob lectins, which contain two or three chitin-binding domains each with six conserved cysteines <cite>Frisardi2000,Van_Dellen2006b</cite>. Mass spectrometry and specific antibodies have confirmed the presence of chitinase, Jessie lectins, and Jacob lectins in cyst walls of ''E. histolytica'', the human pathogen <cite>Ghosh2010,Ali2012</cite>. | |
| − | + | Cyst wall proteins, which are absent from trophozoites but abundant in cysts, coat chitin fibrils and protect them from bacterial chitinases, which are ubiquitous in the environment <cite>Samuelson2011,Ehrenkaufer2013</cite>. One function of CBM55 is to retain chitinase and Jessie 3 lectins in the cyst wall. The chitinase is likely involved in remodeling the cyst wall during encystation and breaking down the cyst wall when the trophozoite escapes during excystation. The daub domain of Jessie 3, which forms a biofilm when produced recombinantly as a fusion with maltose-binding protein in the periplasm of bacteria, makes the cyst wall impenetrable to molecules as small as one kilodalton <cite>Chatterjee2009</cite>. | |
| − | |||
== Family Firsts == | == Family Firsts == | ||
;First Identified | ;First Identified | ||
| − | :A CBM55 was first identified at the N-terminus of the [[GH18]] chitinase of ''Entamoeba histolytica''<cite>Van_Dellen2002, de_la_Vega1997</cite>. | + | :A CBM55 was first identified at the N-terminus of the [[GH18]] chitinase of ''Entamoeba histolytica'' <cite>Van_Dellen2002, de_la_Vega1997</cite>. |
;First Structural Characterization | ;First Structural Characterization | ||
| − | : | + | :There is no structure available. |
== References == | == References == | ||
| Line 48: | Line 48: | ||
#Samuelson2011 pmid=20934911 | #Samuelson2011 pmid=20934911 | ||
#Ehrenkaufer2013 pmid=23889909 | #Ehrenkaufer2013 pmid=23889909 | ||
| + | #Aurrecoechea2011 pmid=20974635 | ||
</biblio> | </biblio> | ||
[[Category:Carbohydrate Binding Module Families|CBM055]] | [[Category:Carbohydrate Binding Module Families|CBM055]] | ||
Latest revision as of 13:15, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM55.html |
Ligand specificities
A CBM55, which is ~60-aa long and contains eight cysteines in conserved positions, was first identified at the N-terminus of the GH18 chitinase of Entamoeba histolytica, the protist that causes dysentery and liver abscess (Figure 1) [1, 2]. CBM55 members are also present at the N-termini of Jessie 3 lectins, which contain a self-aggregating (daub) domain, while CBM55 is the only domain in Jessie 1 and Jessie 2 lectins [3]. CBM55 members are present in GH18 chitinases and Jessie lectins of all five Entamoeba species that have been sequenced [4]. CBM55 members are absent from other eukaryotes, eubacteria, and archaea, and so the CBM55 motif appears to have been “created from scratch” by the common ancestor to Entamoebae. CBM55 members of E. histolytica chitinase, Jessie 1, and Jessie 3 lectins, each produced by gene expression under a constitutive actin promoter in transformed trophozoites, demonstrated binding to chitin beads [1].
Structural Features
While the presence of eight cysteines in conserved positions suggest that CBM55 is a disulfide knot, there is no structure for CBM55 [1]. Because CBM55 binds to chitin fibrils in the cyst wall, it is a type A CBM [3].
Functionalities
In the human intestine, motile E. histolytica trophozoites phagocytose bacteria and/or attach to and take bites out of (trogocytose) host epithelial cells [5]. Alternatively, trophozoites form cysts, which have chitin and deacetylated chitin (chitosan) in their wall [6, 7]. E. histolytica is transmitted when cysts are shed in the feces and contaminate food, soil, or water. When cysts are ingested, trophozoites escape their wall and are released into the intestine, completing the life cycle. The most abundant proteins identified by mass spectrometry of cyst walls of E. invadens, a model for the human pathogen, are chitinases, Jessie 3 lectins, and Jacob lectins, which contain two or three chitin-binding domains each with six conserved cysteines [8, 9]. Mass spectrometry and specific antibodies have confirmed the presence of chitinase, Jessie lectins, and Jacob lectins in cyst walls of E. histolytica, the human pathogen [10, 11]. Cyst wall proteins, which are absent from trophozoites but abundant in cysts, coat chitin fibrils and protect them from bacterial chitinases, which are ubiquitous in the environment [12, 13]. One function of CBM55 is to retain chitinase and Jessie 3 lectins in the cyst wall. The chitinase is likely involved in remodeling the cyst wall during encystation and breaking down the cyst wall when the trophozoite escapes during excystation. The daub domain of Jessie 3, which forms a biofilm when produced recombinantly as a fusion with maltose-binding protein in the periplasm of bacteria, makes the cyst wall impenetrable to molecules as small as one kilodalton [3].
Family Firsts
- First Identified
- A CBM55 was first identified at the N-terminus of the GH18 chitinase of Entamoeba histolytica [1, 2].
- First Structural Characterization
- There is no structure available.
References
- Van Dellen K, Ghosh SK, Robbins PW, Loftus B, and Samuelson J. (2002). Entamoeba histolytica lectins contain unique 6-Cys or 8-Cys chitin-binding domains. Infect Immun. 2002;70(6):3259-63. DOI:10.1128/IAI.70.6.3259-3263.2002 |
- de la Vega H, Specht CA, Semino CE, Robbins PW, Eichinger D, Caplivski D, Ghosh S, and Samuelson J. (1997). Cloning and expression of chitinases of Entamoebae. Mol Biochem Parasitol. 1997;85(2):139-47. DOI:10.1016/s0166-6851(96)02817-4 |
- Chatterjee A, Ghosh SK, Jang K, Bullitt E, Moore L, Robbins PW, and Samuelson J. (2009). Evidence for a "wattle and daub" model of the cyst wall of entamoeba. PLoS Pathog. 2009;5(7):e1000498. DOI:10.1371/journal.ppat.1000498 |
- Aurrecoechea C, Barreto A, Brestelli J, Brunk BP, Caler EV, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Iodice J, Kissinger JC, Kraemer ET, Li W, Nayak V, Pennington C, Pinney DF, Pitts B, Roos DS, Srinivasamoorthy G, Stoeckert CJ Jr, Treatman C, and Wang H. (2011). AmoebaDB and MicrosporidiaDB: functional genomic resources for Amoebozoa and Microsporidia species. Nucleic Acids Res. 2011;39(Database issue):D612-9. DOI:10.1093/nar/gkq1006 |
- Ralston KS, Solga MD, Mackey-Lawrence NM, Somlata, Bhattacharya A, and Petri WA Jr. (2014). Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature. 2014;508(7497):526-30. DOI:10.1038/nature13242 |
- Van Dellen KL, Bulik DA, Specht CA, Robbins PW, and Samuelson JC. (2006). Heterologous expression of an Entamoeba histolytica chitin synthase in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5(1):203-6. DOI:10.1128/EC.5.1.203-206.2006 |
- Das S, Van Dellen K, Bulik D, Magnelli P, Cui J, Head J, Robbins PW, and Samuelson J. (2006). The cyst wall of Entamoeba invadens contains chitosan (deacetylated chitin). Mol Biochem Parasitol. 2006;148(1):86-92. DOI:10.1016/j.molbiopara.2006.03.002 |
- Frisardi M, Ghosh SK, Field J, Van Dellen K, Rogers R, Robbins P, and Samuelson J. (2000). The most abundant glycoprotein of amebic cyst walls (Jacob) is a lectin with five Cys-rich, chitin-binding domains. Infect Immun. 2000;68(7):4217-24. DOI:10.1128/IAI.68.7.4217-4224.2000 |
- Van Dellen KL, Chatterjee A, Ratner DM, Magnelli PE, Cipollo JF, Steffen M, Robbins PW, and Samuelson J. (2006). Unique posttranslational modifications of chitin-binding lectins of Entamoeba invadens cyst walls. Eukaryot Cell. 2006;5(5):836-48. DOI:10.1128/EC.5.5.836-848.2006 |
- Ghosh SK, Van Dellen KL, Chatterjee A, Dey T, Haque R, Robbins PW, and Samuelson J. (2010). The Jacob2 lectin of the Entamoeba histolytica cyst wall binds chitin and is polymorphic. PLoS Negl Trop Dis. 2010;4(7):e750. DOI:10.1371/journal.pntd.0000750 |
- Ali IK, Haque R, Siddique A, Kabir M, Sherman NE, Gray SA, Cangelosi GA, and Petri WA Jr. (2012). Proteomic analysis of the cyst stage of Entamoeba histolytica. PLoS Negl Trop Dis. 2012;6(5):e1643. DOI:10.1371/journal.pntd.0001643 |
- Samuelson J and Robbins P. (2011). A simple fibril and lectin model for cyst walls of Entamoeba and perhaps Giardia. Trends Parasitol. 2011;27(1):17-22. DOI:10.1016/j.pt.2010.09.002 |
- Ehrenkaufer GM, Weedall GD, Williams D, Lorenzi HA, Caler E, Hall N, and Singh U. (2013). The genome and transcriptome of the enteric parasite Entamoeba invadens, a model for encystation. Genome Biol. 2013;14(7):R77. DOI:10.1186/gb-2013-14-7-r77 |