CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Carbohydrate Esterase Family 20"
| Line 31: | Line 31: | ||
Carbohydrate esterase family 20 (CE20) currently comprises xyloglucan acetylesterases (E.C. [https://www.enzyme-database.org/query.php?ec=3.1.1.6 3.2.1.6]) <cite>Vieira2021</cite> and acetylesterases putatively related to arabinoxylan deacetylation <cite>Liu2022</cite>. | Carbohydrate esterase family 20 (CE20) currently comprises xyloglucan acetylesterases (E.C. [https://www.enzyme-database.org/query.php?ec=3.1.1.6 3.2.1.6]) <cite>Vieira2021</cite> and acetylesterases putatively related to arabinoxylan deacetylation <cite>Liu2022</cite>. | ||
| − | [[Image:Ce20_cazypedia.png|thumb|500px|Figure 1: The tridimensional structure and architecture of ''Xac''XaeA, the founding member of CE20 family. [https://www.rcsb.org/structure/7KMM PDB ID 7KMM] (a) Domain organization showing the position of catalytic residues (red triangles). (b) Structural topology, delimited by domains and (c) crystal structure, color-coded according to (a). Catalytic residues are circled. X448 domain is absent in the tridimensional model due to proteolysis removal in the crystallization steps. Adapted from <cite>Vieira2021</cite> originally published under [http://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License].]]== Catalytic | + | [[Image:Ce20_cazypedia.png|thumb|500px|Figure 1: The tridimensional structure and architecture of ''Xac''XaeA, the founding member of CE20 family. [https://www.rcsb.org/structure/7KMM PDB ID 7KMM] (a) Domain organization showing the position of catalytic residues (red triangles). (b) Structural topology, delimited by domains and (c) crystal structure, color-coded according to (a). Catalytic residues are circled. X448 domain is absent in the tridimensional model due to proteolysis removal in the crystallization steps. Adapted from <cite>Vieira2021</cite> originally published under [http://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License].]]== Catalytic residues == |
''Xac''XaeA, the founding member of family CE20 <cite>Vieira2021</cite>, harbors the canonical catalytic triad Ser-Asp-His and the conserved electropositive oxyanion hole. Thus, it is supposed to have the same mechanism of action described for other carbohydrate esterases, such as the members of family [[CE3]] and [[CE6]]. However, this assumption still requires experimental validation. The catalytic-relevant residues are present in consensus regions in ''Xanthomonas'', denoted as Block I (GQ'''S'''NME) and Block V ('''D'''I'''H''') (Infered catalytic residues in bold). Oxyanion hole residues are present in Blocks I (GQSNME) and III (YQGET). Although consensus Block II is present, Block IV is absent <cite>Vieira2021</cite>, as noted for other related carbohydrate esterase (CE) families. | ''Xac''XaeA, the founding member of family CE20 <cite>Vieira2021</cite>, harbors the canonical catalytic triad Ser-Asp-His and the conserved electropositive oxyanion hole. Thus, it is supposed to have the same mechanism of action described for other carbohydrate esterases, such as the members of family [[CE3]] and [[CE6]]. However, this assumption still requires experimental validation. The catalytic-relevant residues are present in consensus regions in ''Xanthomonas'', denoted as Block I (GQ'''S'''NME) and Block V ('''D'''I'''H''') (Infered catalytic residues in bold). Oxyanion hole residues are present in Blocks I (GQSNME) and III (YQGET). Although consensus Block II is present, Block IV is absent <cite>Vieira2021</cite>, as noted for other related carbohydrate esterase (CE) families. | ||
Revision as of 11:32, 22 March 2023
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
| Carbohydrate Esterase Family CE20 | |
| Fold | β-sandwich/(α-β-α) sandwich/β-sandwich |
| Mechanism | serine hydrolase |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/CE20.html | |
Substrate specificities
Carbohydrate esterase family 20 (CE20) currently comprises xyloglucan acetylesterases (E.C. 3.2.1.6) [1] and acetylesterases putatively related to arabinoxylan deacetylation [2].
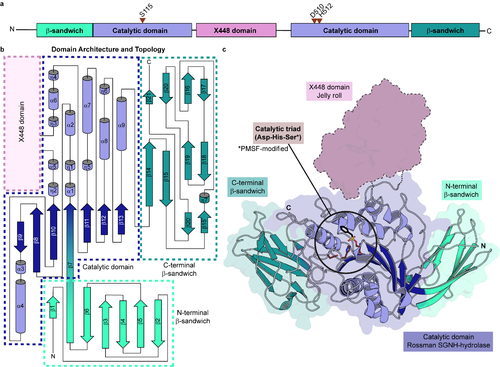
== Catalytic residues ==
XacXaeA, the founding member of family CE20 [1], harbors the canonical catalytic triad Ser-Asp-His and the conserved electropositive oxyanion hole. Thus, it is supposed to have the same mechanism of action described for other carbohydrate esterases, such as the members of family CE3 and CE6. However, this assumption still requires experimental validation. The catalytic-relevant residues are present in consensus regions in Xanthomonas, denoted as Block I (GQSNME) and Block V (DIH) (Infered catalytic residues in bold). Oxyanion hole residues are present in Blocks I (GQSNME) and III (YQGET). Although consensus Block II is present, Block IV is absent [1], as noted for other related carbohydrate esterase (CE) families.
Kinetics and Mechanism
XacXaeA is specific to O-acetylation since it is not capable of cleaving N-acetylated carbohydrates. It shows activity on a broad range of O-acetylated mono- and disaccharides and did not show a positional preference for acetylated oxygens. XacXaeA was active towards cell wall extracted xyloglucan oligosaccharides, deacetylating distinct types of structures such as XXLG/XLXG, XXFG, and XLFG [1]. Kinetic data for the second characterized member of family CE20, XuaJ, is available for the substrate 1-Naphthyl acetate [2].
Three-dimensional structures
The CE20 structure is composed of a central catalytic core, which displays the SGNH hydrolase fold, flanked by two antiparallel seven-stranded β-sandwiches intimately linked to the central core, forming a monolithic structure [1]. Such structural architecture diverges from CE families described in the CAZy database so far. In XacXaeA, the founding member of family CE20 [1], the catalytic core is composed of two halves (residues 104-216 and 397-541) due to the insertion of a domain (residues 217-396, named X448 in the CAZy database [3, 4]) in the α5-η3 loop.
Family Firsts
- First stereochemistry determination
- Content is to be added here.
- First catalytic nucleophile identification
- Content is to be added here.
- First general acid/base residue identification
- Content is to be added here.
- First biochemical characterization
- Xanthomonas citri subsp. citri 306 xyloglucan acetylesterase in 2021 [1].
- First 3-D structure
- Xanthomonas citri subsp. citri 306 xyloglucan acetylesterase crystal structure in 2021 PDB ID 7KMM [1].
References
Error fetching PMID 36403068:
- Error fetching PMID 34193873:
- Error fetching PMID 36403068:
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, and Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233-8. DOI:10.1093/nar/gkn663 |
-
Davies, G.J. and Sinnott, M.L. (2008) Sorting the diverse: the sequence-based classifications of carbohydrate-active enzymes. The Biochemist, vol. 30, no. 4., pp. 26-32. Download PDF version.