CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Carbohydrate Binding Module Family 101"
(Created page with " <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> {{UnderConstruc...") |
|||
| (12 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
* [[Author]]: [[User:Xuanwei Mei|Xuanwei Mei]] | * [[Author]]: [[User:Xuanwei Mei|Xuanwei Mei]] | ||
* [[Responsible Curator]]: [[User:Yaoguang Chang|Yaoguang Chang]] | * [[Responsible Curator]]: [[User:Yaoguang Chang|Yaoguang Chang]] | ||
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | |||
| − | '' | + | [[File:Figure.png|thumb|'''Figure 1. Carbohydrate array assays of WfCBM101. ''' The binding of WfCBM101 to agarose and other galactans in red algae (A), and several other polysaccharides (B) was evaluated. κ-Car, κ-carrageenan. ι-Car, ι-carrageenan. Alg, alginate. An-FUC, sulfated fucan from ''Ascophyllum nodosum''. CSA, chondroitin sulfate A sodium salt. HA, hyaluronic acid. Pec, pectin. Three repeats were conducted for each polysaccharide.''' ]] |
| + | |||
| + | The first characterized member in the CBM101 family is WfCBM101 <cite>Mei2023</cite>. The CBM WfCBM101 bound to agarose and displayed a weak affinity to porphyran (Fig. 1). It was incapable of binding to the other polysaccharides that were examined, including κ-carrageenan, ι-carrageenan, alginate, sulfated fucan, chondroitin sulfate A sodium salt, hyaluronic acid, and pectin. Furthermore, WfCBM101 bound to agarose tetrasaccharide, but not to porphyran tetrasaccharide. It was reported porphyran consists of approximately 30% structural units of agarose <cite>Chi2012</cite>. It is thus speculated that the weak affinity of WfCBM101 for porphyran is attributed to the structural heterogeneity of porphyran. | ||
== Structural Features == | == Structural Features == | ||
| − | + | An AlphaFold2 model predicts that WfCBM101 has a β-sandwich fold. | |
| − | |||
| − | |||
| − | |||
== Functionalities == | == Functionalities == | ||
| − | '' | + | To evaluate the feasibility of WfCBM101 as a tool in the ''in situ'' investigation of porphyran, a fluorescent probe was constructed by fusing WfCBM101 with a green fluorescent protein. The ''in situ'' visualization of agarose in the red alga ''Gelidium amansii'' was realized by utilizing the fluorescent probe <cite>Mei2023</cite>. |
| − | + | Taking WfCBM101 as the query sequence, 15 modules were retrieved from the NCBI database by the BLASTP program (E-value < e<sup>-5</sup>). There are eight modules adjacent to catalytic domains which are divided into the [[GH16]]_16 subfamily or [[GH86]] family. According to the CAZy database, the [[GH16]_16 subfamily and [[GH86]] family have members that degrade agarose. This implies that these eight modules might bind to agarose; however, this requires further investigation. | |
| − | |||
| − | |||
== Family Firsts == | == Family Firsts == | ||
;First Identified | ;First Identified | ||
| − | + | The first member WfCBM101 is a component of a GH86 β-agarase (unpublished data) from a marine bacterium ''Wenyingzhuangia fucanilytica'' CZ1127<sup>T</sup> <cite>Chen2016</cite>. | |
;First Structural Characterization | ;First Structural Characterization | ||
| − | + | No experimentally determined three-dimensional structure has been solved in this CBM family. | |
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Mei2023 pmid=38010608 |
| − | # | + | #Chi2012 pmid=22526785 |
| − | + | #Chen2016 pmid=27220912 | |
| − | |||
| − | |||
| − | |||
| − | # | ||
</biblio> | </biblio> | ||
<!-- Do not delete this Category tag --> | <!-- Do not delete this Category tag --> | ||
[[Category:Carbohydrate Binding Module Families|CBM101]] | [[Category:Carbohydrate Binding Module Families|CBM101]] | ||
| − | |||
Latest revision as of 00:56, 16 February 2024
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| http://www.cazy.org/CBM101.html |
Ligand specificities
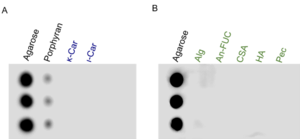
The first characterized member in the CBM101 family is WfCBM101 [1]. The CBM WfCBM101 bound to agarose and displayed a weak affinity to porphyran (Fig. 1). It was incapable of binding to the other polysaccharides that were examined, including κ-carrageenan, ι-carrageenan, alginate, sulfated fucan, chondroitin sulfate A sodium salt, hyaluronic acid, and pectin. Furthermore, WfCBM101 bound to agarose tetrasaccharide, but not to porphyran tetrasaccharide. It was reported porphyran consists of approximately 30% structural units of agarose [2]. It is thus speculated that the weak affinity of WfCBM101 for porphyran is attributed to the structural heterogeneity of porphyran.
Structural Features
An AlphaFold2 model predicts that WfCBM101 has a β-sandwich fold.
Functionalities
To evaluate the feasibility of WfCBM101 as a tool in the in situ investigation of porphyran, a fluorescent probe was constructed by fusing WfCBM101 with a green fluorescent protein. The in situ visualization of agarose in the red alga Gelidium amansii was realized by utilizing the fluorescent probe [1]. Taking WfCBM101 as the query sequence, 15 modules were retrieved from the NCBI database by the BLASTP program (E-value < e-5). There are eight modules adjacent to catalytic domains which are divided into the GH16_16 subfamily or GH86 family. According to the CAZy database, the [[GH16]_16 subfamily and GH86 family have members that degrade agarose. This implies that these eight modules might bind to agarose; however, this requires further investigation.
Family Firsts
- First Identified
The first member WfCBM101 is a component of a GH86 β-agarase (unpublished data) from a marine bacterium Wenyingzhuangia fucanilytica CZ1127T [3].
- First Structural Characterization
No experimentally determined three-dimensional structure has been solved in this CBM family.
References
- Mei X, Zhang Y, Jiang X, Liu G, Shen J, Xue C, Xiao H, and Chang Y. (2024). Discovery and characterization of a novel carbohydrate-binding module: a favorable tool for investigating agarose. J Sci Food Agric. 2024;104(5):2792-2797. DOI:10.1002/jsfa.13164 |
- Chi WJ, Chang YK, and Hong SK. (2012). Agar degradation by microorganisms and agar-degrading enzymes. Appl Microbiol Biotechnol. 2012;94(4):917-30. DOI:10.1007/s00253-012-4023-2 |
- Chen F, Chang Y, Dong S, and Xue C. (2016). Wenyingzhuangia fucanilytica sp. nov., a sulfated fucan utilizing bacterium isolated from shallow coastal seawater. Int J Syst Evol Microbiol. 2016;66(9):3270-3275. DOI:10.1099/ijsem.0.001184 |