CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 168"
Harry Brumer (talk | contribs) (Created page with "{{UnassignedPage}} GH168") |
Harry Brumer (talk | contribs) |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{ | + | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> |
| + | {{CuratorApproved}} | ||
| + | * [[Author]]: [[User:Jingjing Shen|Jingjing Shen]] | ||
| + | * [[Responsible Curator]]: [[User:Yaoguang Chang|Yaoguang Chang]] | ||
| + | ---- | ||
| + | <!-- The data in the table below should be updated by the Author/Curator according to current information on the family --> | ||
| + | <div style="float:right"> | ||
| + | {| {{Prettytable}} | ||
| + | |- | ||
| + | |{{Hl2}} colspan="2" align="center" |'''Glycoside Hydrolase Family GH168''' | ||
| + | |- | ||
| + | |'''Clan''' | ||
| + | |GH-x | ||
| + | |- | ||
| + | |'''Mechanism''' | ||
| + | |retaining | ||
| + | |- | ||
| + | |'''Active site residues''' | ||
| + | |not known | ||
| + | |- | ||
| + | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
| + | |- | ||
| + | | colspan="2" |{{CAZyDBlink}}GH168.html | ||
| + | |} | ||
| + | </div> | ||
| + | <!-- This is the end of the table --> | ||
| + | |||
| + | |||
| + | == Substrate specificities == | ||
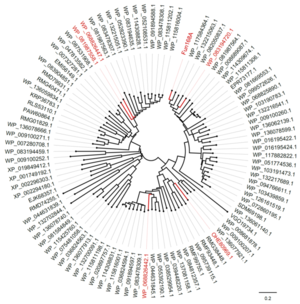
| + | Members of GH168 have been shown to exhibit α-1,3-L-fucanase activity. The first member of this family, Fun168A from a marine bacterium ''Wenyingzhuangia funcanilytica'' CZ1127<sup>T</sup>, specifically hydrolyzes the α-1,3- L-fucoside bonds between 2-O-sulfated and non-sulfated fucose residues in the sulfated fucan from sea cucumber ''Isostichopus badionotus'' in a random endo-acting manner. Meanwhile, [http://www.cazy.org/GH168_characterized.html five homologues of Fun168A display activities toward sulfated fucan] from ''Isostichopus badionotus'' <cite>Shen2020</cite>. | ||
| + | [[File:Tree of GH168.png|thumb|'''Figure 1. The phylogenetic tree of GH168 homologues.''' Sequences comfirmed to exhibit α-1,3-L-fucanase activity were in red.]] | ||
| + | |||
| + | == Kinetics and Mechanism == | ||
| + | ''W. funcanilytica'' Fun168A exhibited transglycosylating activity with glycerin, methanol, and L-fucose as acceptors. This transglycosylating activity implied a [[retaining]] mechanism of catalysis <cite>Shen2020</cite>. | ||
| + | |||
| + | == Catalytic Residues == | ||
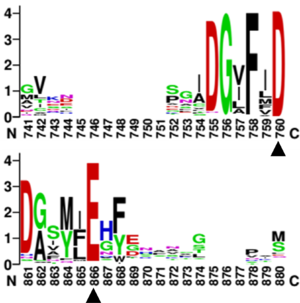
| + | Multiple sequence alignments of GH168 homologues showed that D206 and E264 in Fun168A were strictly conserved in all sequences. Two single-site mutants of ''W. funcanilytica'' Fun168A, D206E and E264Q, were inactive on Ib-FUC, indicating that D206 and E264 were critical for activity <cite>Shen2020</cite>. | ||
| + | [[File:catalytic residues.png|thumb|'''Figure 2. Multiple sequence alignments of residues in GH168 homologues.''' The strictly conserved residues in all sequences are indicated with black triangles.]] | ||
| + | |||
| + | == Three-dimensional structures == | ||
| + | No three-dimensional structure has been solved in this family at present. | ||
| + | |||
| + | == Family Firsts == | ||
| + | ;First stereochemistry determination: Not yet identified. | ||
| + | ;First catalytic nucleophile identification: Not yet identified. | ||
| + | ;First general acid/base residue identification: Not yet identified. | ||
| + | ;First 3-D structure: Not yet identified. | ||
| + | |||
| + | == References == | ||
| + | <biblio> | ||
| + | #Shen2020 pmid=32849348 | ||
| + | </biblio> | ||
| + | |||
| + | <!-- Do not delete this Category tag --> | ||
[[Category:Glycoside Hydrolase Families|GH168]] | [[Category:Glycoside Hydrolase Families|GH168]] | ||
Latest revision as of 12:23, 18 June 2023
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH168 | |
| Clan | GH-x |
| Mechanism | retaining |
| Active site residues | not known |
| CAZy DB link | |
| http://www.cazy.org/GH168.html | |
Substrate specificities
Members of GH168 have been shown to exhibit α-1,3-L-fucanase activity. The first member of this family, Fun168A from a marine bacterium Wenyingzhuangia funcanilytica CZ1127T, specifically hydrolyzes the α-1,3- L-fucoside bonds between 2-O-sulfated and non-sulfated fucose residues in the sulfated fucan from sea cucumber Isostichopus badionotus in a random endo-acting manner. Meanwhile, five homologues of Fun168A display activities toward sulfated fucan from Isostichopus badionotus [1].
Kinetics and Mechanism
W. funcanilytica Fun168A exhibited transglycosylating activity with glycerin, methanol, and L-fucose as acceptors. This transglycosylating activity implied a retaining mechanism of catalysis [1].
Catalytic Residues
Multiple sequence alignments of GH168 homologues showed that D206 and E264 in Fun168A were strictly conserved in all sequences. Two single-site mutants of W. funcanilytica Fun168A, D206E and E264Q, were inactive on Ib-FUC, indicating that D206 and E264 were critical for activity [1].
Three-dimensional structures
No three-dimensional structure has been solved in this family at present.
Family Firsts
- First stereochemistry determination
- Not yet identified.
- First catalytic nucleophile identification
- Not yet identified.
- First general acid/base residue identification
- Not yet identified.
- First 3-D structure
- Not yet identified.