CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Polysaccharide Lyase Family 6"
Emil Stender (talk | contribs) |
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
||
| (6 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
* [[Author]]: [[User:Emil Stender|Emil G.P. Stender]] | * [[Author]]: [[User:Emil Stender|Emil G.P. Stender]] | ||
| − | * [[Responsible Curator]]: | + | * [[Responsible Curator]]: [[User:Birte Svensson|Birte Svensson]] |
---- | ---- | ||
| Line 32: | Line 32: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | PL6 contains 3 subfamilies <cite>Lombard2010</cite> all of which contain members catalyzing the depolymerisation of alginate <cite>Mathieu2016</cite>. Alginate consist of 1,4 linked β-D-mannuronic acid and α-L-guluronic acid arranged in poly-mannuronic acid blocks, poly-guluronic acid blocks or poly-mannuronic/guluronic acid blocks <cite>Haug1966 Haug1967</cite>. Subfamily 2 and 3 have so far only shown specificity for poly-mannuronic/guluronic acid blocks <cite>Mathieu2016</cite>, while subfamily 1 has been demonstrated to depolymerize poly-guluronic acid <cite>Lyu2019 Xu2017</cite>, poly-mannuronic acid <cite>Maki1993 Stender2019</cite>, poly-mannuronic/guluronic acid <cite>Mathieu2016</cite> as well as dermatan sulfate (formerly chrondroitin B) <cite>Mathieu2016 #Huang1999 #Michel2004</cite>. Dermatan sulfate | + | PL6 contains 3 subfamilies <cite>Lombard2010 Mathieu2016</cite> all of which contain members catalyzing the depolymerisation of alginate <cite>Mathieu2016</cite>. Alginate consist of 1,4 linked β-{{Smallcaps|D}}-mannuronic acid and α-{{Smallcaps|L}}-guluronic acid arranged in poly-mannuronic acid blocks, poly-guluronic acid blocks or poly-mannuronic/guluronic acid blocks <cite>Haug1966 Haug1967</cite>. Subfamily 2 and 3 have so far only shown specificity for poly-mannuronic/guluronic acid blocks <cite>Mathieu2016</cite>, while subfamily 1 has been demonstrated to depolymerize poly-guluronic acid <cite>Lyu2019 Xu2017</cite>, poly-mannuronic acid <cite>Maki1993 Stender2019</cite>, poly-mannuronic/guluronic acid <cite>Mathieu2016</cite> as well as dermatan sulfate (formerly chrondroitin B) <cite>Mathieu2016 #Huang1999 #Michel2004</cite>. Dermatan sulfate consist of N-acetyl galactosamine (GalNAc) and glucuronic acid (GlcA) joined by β 1,4 or 1,3 linkages respectively with a variable sulfation pattern <cite>Trowbridge2002</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | [[Image:PL6_lyase_mechanism.png|thumb|600px| '''Figure 1.''' ''Syn'' – or ''anti'' – β-elimination catalyzed by PL6 enzymes acting on alginate. M represents mannuronic acid and G guluronic acid. n represents the continued sugar chain.]] | + | [[Image:PL6_lyase_mechanism.png|thumb|600px| '''Figure 1.''' ''Syn'' – or ''anti'' – β-elimination catalyzed by PL6 enzymes acting on alginate. M represents mannuronic acid and G guluronic acid. n represents the continued sugar chain. In both cases the catalytic base abstracts the C5 proton and an acid donates one resulting in the β-elimination of the 1,4 glycosidic linkage.]] |
| − | The β-elimination catalyzed by the PL6 enzymes results in the formation of a C4-C5 unsaturated sugar at the new non-reducing end. The first step is the neutralization of the acid group in the +1 subsite by a calcium <cite>Xu2017 Michel2004</cite> or by water <cite>Lyu2019</cite>. This lowers the | + | The β-elimination catalyzed by the PL6 enzymes results in the formation of a C4-C5 unsaturated sugar residue at the new non-reducing end. The first step is the neutralization of the acid group in the +1 subsite by a calcium <cite>Xu2017 Michel2004</cite> or by water <cite>Lyu2019</cite>. This lowers the p''K''<sub>a</sub> value of the C5-proton allowing for abstraction by the catalytic base (Figure 1). A catalytic acid then donates a proton to the glycosidic linkage resulting in the β-elimination. This can be done in ''syn'' with the acid and base on the same side of the sugar ring in the transition state (the case for {{Smallcaps|D}}-mannuronic acid) or ''anti'' where they are on opposite sides of the sugar ring (the case for {{Smallcaps|L}}-guluronic acid) <cite>Garron2010 Xu2018</cite>. |
== Catalytic Residues == | == Catalytic Residues == | ||
| − | After charge neutralization a lysine functions as the catalytic base and an arginine the acid. They were originally identified as K253 and R273 in chondroitinase B from ''Pedobacter | + | After charge neutralization a lysine functions as the catalytic base and an arginine as the acid. They were originally identified as K253 and R273 in chondroitinase B from ''Pedobacter heparinus'' <cite>Huang1999</cite>. PL6 is so far the only discovered alginate lyase family that uses K/R as a catalytic base/acid pair <cite>Xu2018</cite>. |
== Three-dimensional structures == | == Three-dimensional structures == | ||
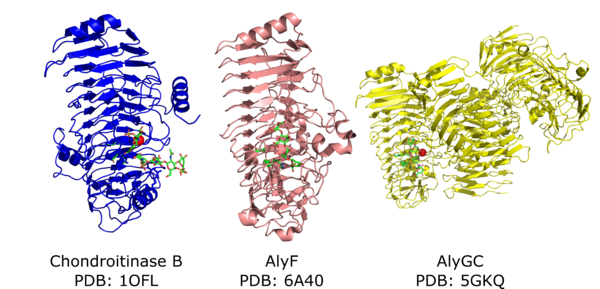
| − | [[Image:PL6_structures.png|thumb|600px|'''Figure 2.''' Crystal structures of the monomeric chrondroitinase B and AlyF | + | [[Image:PL6_structures.png|thumb|600px|'''Figure 2.''' Crystal structures of the monomeric chrondroitinase B and AlyF as well as the dimeric AlyGC. Catalytic residues and substrates are in green, the neutralizing calcium is the red sphere.]] |
| − | PL6 catalytic domain adopts a parallel β-helix fold with the | + | PL6 catalytic domain adopts a parallel β-helix fold with the active site located on the surface of one of the β-sheets (Figure 2). The first PL6 structure solved was the chrondoitinase B from ''Pedobacter heparinus'' (1.7 Å) <cite>Huang1999</cite> later it was shown that this enzyme is calcium dependent <cite>Michel2004</cite>. The first alginate lyase structure solved was the exolytic, guluronic acid-specific, homo-dimeric AlyGC in complex with tetra-mannuronic acid (2.6 Å) <cite>Xu2017</cite>. The first monomeric alginate lyase structure solved was the guluronic acid-specific AlyF in complex with tetra-guluronic acid (1.8 Å) <cite>Lyu2019</cite>. The first mannuronic acid specific alginate lyase structure was ''Bcel''PL6 (1.3Å) from human gut ''Bacteroides cellulosilyticus'' <cite>Stender2019</cite>. All four structures belong to subfamily 1. There are no available crystal structures from subfamilies 2 and 3. |
== Family Firsts == | == Family Firsts == | ||
;First catalytic activity: OS-ALG-9 from ''Pseudomonas'' sp. on non-purified recombinant enzyme by the thiobarbituric acid method <cite>Maki1993</cite> | ;First catalytic activity: OS-ALG-9 from ''Pseudomonas'' sp. on non-purified recombinant enzyme by the thiobarbituric acid method <cite>Maki1993</cite> | ||
| − | ;First catalytic base/acid: The catalytic arginine was originally identified in Chondroitinase B from ''Pedobacter | + | ;First catalytic base/acid: The catalytic arginine was originally identified in Chondroitinase B from ''Pedobacter heparinus'' based on the crystal structure, concervation, mutagenesis and activity analysis (R271E no activity, R271K 0.09 % activity) <cite>Michel2004</cite>. The catalytic lysine was identified later based on conservation, mutagenesis and activity analysis <cite>Xu2017</cite> |
;First charge neutralizer: Calcium in Chondroitinase B from ''Pedobacter Heparinus'' by by crystallography and assaying the effect of calcium on enzyme activity <cite>Michel2004</cite>. | ;First charge neutralizer: Calcium in Chondroitinase B from ''Pedobacter Heparinus'' by by crystallography and assaying the effect of calcium on enzyme activity <cite>Michel2004</cite>. | ||
;First 3-D structure: Chondroitinase B from ''Pedobacter Heparinus'' <cite>Huang1999</cite>. | ;First 3-D structure: Chondroitinase B from ''Pedobacter Heparinus'' <cite>Huang1999</cite>. | ||
Latest revision as of 14:16, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Polysaccharide Lyase Family 6 | |
| 3D structure | parallel β-helix |
| Mechanism | β-elimination |
| Charge neutralizer | calcium or water |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/PL6.html | |
Substrate specificities
PL6 contains 3 subfamilies [1, 2] all of which contain members catalyzing the depolymerisation of alginate [2]. Alginate consist of 1,4 linked β-D-mannuronic acid and α-L-guluronic acid arranged in poly-mannuronic acid blocks, poly-guluronic acid blocks or poly-mannuronic/guluronic acid blocks [3, 4]. Subfamily 2 and 3 have so far only shown specificity for poly-mannuronic/guluronic acid blocks [2], while subfamily 1 has been demonstrated to depolymerize poly-guluronic acid [5, 6], poly-mannuronic acid [7, 8], poly-mannuronic/guluronic acid [2] as well as dermatan sulfate (formerly chrondroitin B) [2, 9, 10]. Dermatan sulfate consist of N-acetyl galactosamine (GalNAc) and glucuronic acid (GlcA) joined by β 1,4 or 1,3 linkages respectively with a variable sulfation pattern [11].
Kinetics and Mechanism
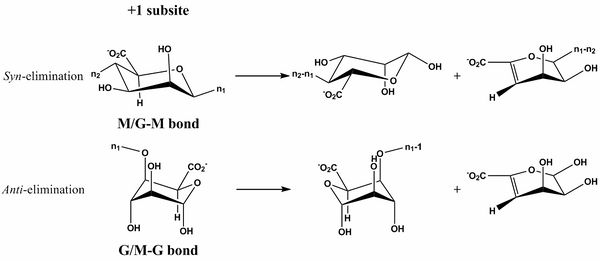
The β-elimination catalyzed by the PL6 enzymes results in the formation of a C4-C5 unsaturated sugar residue at the new non-reducing end. The first step is the neutralization of the acid group in the +1 subsite by a calcium [6, 10] or by water [5]. This lowers the pKa value of the C5-proton allowing for abstraction by the catalytic base (Figure 1). A catalytic acid then donates a proton to the glycosidic linkage resulting in the β-elimination. This can be done in syn with the acid and base on the same side of the sugar ring in the transition state (the case for D-mannuronic acid) or anti where they are on opposite sides of the sugar ring (the case for L-guluronic acid) [12, 13].
Catalytic Residues
After charge neutralization a lysine functions as the catalytic base and an arginine as the acid. They were originally identified as K253 and R273 in chondroitinase B from Pedobacter heparinus [9]. PL6 is so far the only discovered alginate lyase family that uses K/R as a catalytic base/acid pair [13].
Three-dimensional structures
PL6 catalytic domain adopts a parallel β-helix fold with the active site located on the surface of one of the β-sheets (Figure 2). The first PL6 structure solved was the chrondoitinase B from Pedobacter heparinus (1.7 Å) [9] later it was shown that this enzyme is calcium dependent [10]. The first alginate lyase structure solved was the exolytic, guluronic acid-specific, homo-dimeric AlyGC in complex with tetra-mannuronic acid (2.6 Å) [6]. The first monomeric alginate lyase structure solved was the guluronic acid-specific AlyF in complex with tetra-guluronic acid (1.8 Å) [5]. The first mannuronic acid specific alginate lyase structure was BcelPL6 (1.3Å) from human gut Bacteroides cellulosilyticus [8]. All four structures belong to subfamily 1. There are no available crystal structures from subfamilies 2 and 3.
Family Firsts
- First catalytic activity
- OS-ALG-9 from Pseudomonas sp. on non-purified recombinant enzyme by the thiobarbituric acid method [7]
- First catalytic base/acid
- The catalytic arginine was originally identified in Chondroitinase B from Pedobacter heparinus based on the crystal structure, concervation, mutagenesis and activity analysis (R271E no activity, R271K 0.09 % activity) [10]. The catalytic lysine was identified later based on conservation, mutagenesis and activity analysis [6]
- First charge neutralizer
- Calcium in Chondroitinase B from Pedobacter Heparinus by by crystallography and assaying the effect of calcium on enzyme activity [10].
- First 3-D structure
- Chondroitinase B from Pedobacter Heparinus [9].
References
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, and Henrissat B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432(3):437-44. DOI:10.1042/BJ20101185 |
- Mathieu S, Henrissat B, Labre F, Skjåk-Bræk G, and Helbert W. (2016). Functional Exploration of the Polysaccharide Lyase Family PL6. PLoS One. 2016;11(7):e0159415. DOI:10.1371/journal.pone.0159415 |
-
Haug, A., Larsen, B., and Smidsrod, O. (1966) A study of constitution of alginic acid by partial acid hydrolysis. Acta Chem. Scand. 20, 183–190. DOI:10.3891/acta.chem.scand.20-0183
-
Haug, A., Larsen, B., and Smidsrod, O. (1967) Studies on sequence of uronic acid residues in alginic acid. Acta Chem. Scand. 21, 691–704. DOI:10.3891/acta.chem.scand.21-0691
- Lyu Q, Zhang K, Shi Y, Li W, Diao X, and Liu W. (2019). Structural insights into a novel Ca(2+)-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution. Biochim Biophys Acta Gen Subj. 2019;1863(7):1167-1176. DOI:10.1016/j.bbagen.2019.04.013 |
- Xu F, Dong F, Wang P, Cao HY, Li CY, Li PY, Pang XH, Zhang YZ, and Chen XL. (2017). Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6. J Biol Chem. 2017;292(11):4457-4468. DOI:10.1074/jbc.M116.766030 |
- Maki H, Mori A, Fujiyama K, Kinoshita S, and Yoshida T. (1993). Cloning, sequence analysis and expression in Escherichia coli of a gene encoding an alginate lyase from Pseudomonas sp. OS-ALG-9. J Gen Microbiol. 1993;139(5):987-93. DOI:10.1099/00221287-139-5-987 |
- Stender EGP, Dybdahl Andersen C, Fredslund F, Holck J, Solberg A, Teze D, Peters GHJ, Christensen BE, Aachmann FL, Welner DH, and Svensson B. (2019). Structural and functional aspects of mannuronic acid-specific PL6 alginate lyase from the human gut microbe Bacteroides cellulosilyticus. J Biol Chem. 2019;294(47):17915-17930. DOI:10.1074/jbc.RA119.010206 |
- Huang W, Matte A, Li Y, Kim YS, Linhardt RJ, Su H, and Cygler M. (1999). Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution. J Mol Biol. 1999;294(5):1257-69. DOI:10.1006/jmbi.1999.3292 |
- Michel G, Pojasek K, Li Y, Sulea T, Linhardt RJ, Raman R, Prabhakar V, Sasisekharan R, and Cygler M. (2004). The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J Biol Chem. 2004;279(31):32882-96. DOI:10.1074/jbc.M403421200 |
- Trowbridge JM and Gallo RL. (2002). Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology. 2002;12(9):117R-25R. DOI:10.1093/glycob/cwf066 |
- Garron ML and Cygler M. (2010). Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20(12):1547-73. DOI:10.1093/glycob/cwq122 |
- Xu F, Wang P, Zhang YZ, and Chen XL. (2018). Diversity of Three-Dimensional Structures and Catalytic Mechanisms of Alginate Lyases. Appl Environ Microbiol. 2018;84(3). DOI:10.1128/AEM.02040-17 |